Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
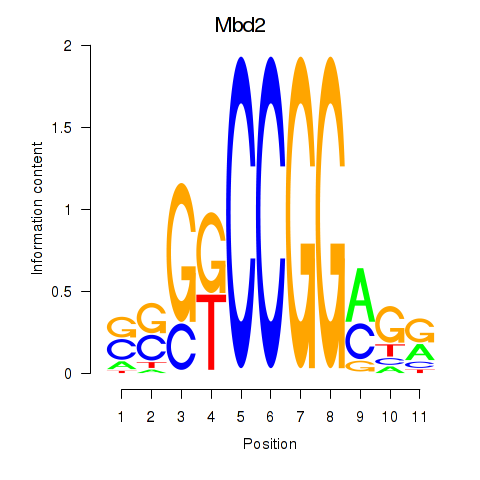
Results for Mbd2
Z-value: 0.47
Transcription factors associated with Mbd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mbd2
|
ENSRNOG00000011853 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mbd2 | rn6_v1_chr18_+_65814026_65814026 | 0.33 | 1.2e-09 | Click! |
Activity profile of Mbd2 motif
Sorted Z-values of Mbd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_172459165 | 15.50 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr5_+_148193710 | 14.31 |
ENSRNOT00000088568
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr9_-_19372673 | 6.96 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr14_-_82048251 | 6.34 |
ENSRNOT00000074734
|
Nat8l
|
N-acetyltransferase 8-like |
| chr6_-_102196138 | 6.04 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr15_+_83707735 | 5.97 |
ENSRNOT00000057843
|
Klf5
|
Kruppel-like factor 5 |
| chr9_+_99998275 | 5.90 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chrX_-_15428518 | 5.84 |
ENSRNOT00000075774
|
LOC108348172
|
proSAAS |
| chr3_+_7109920 | 5.74 |
ENSRNOT00000084092
ENSRNOT00000013809 |
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr7_-_139483997 | 5.58 |
ENSRNOT00000086062
|
Col2a1
|
collagen type II alpha 1 chain |
| chr10_+_62674561 | 5.51 |
ENSRNOT00000019946
ENSRNOT00000056110 |
Ankrd13b
|
ankyrin repeat domain 13B |
| chrX_-_15327684 | 5.49 |
ENSRNOT00000009794
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr18_-_77322690 | 5.44 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr5_-_144779212 | 5.07 |
ENSRNOT00000016230
|
Ncdn
|
neurochondrin |
| chr9_+_82571269 | 5.01 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr12_+_23473270 | 4.81 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr3_-_176465162 | 4.62 |
ENSRNOT00000048807
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr6_-_131926272 | 4.54 |
ENSRNOT00000084057
ENSRNOT00000088421 |
Bcl11b
|
B-cell CLL/lymphoma 11B |
| chr10_+_91254058 | 4.37 |
ENSRNOT00000087218
ENSRNOT00000065373 |
Fmnl1
|
formin-like 1 |
| chr14_-_78902063 | 4.18 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr7_-_141093924 | 4.11 |
ENSRNOT00000086276
|
Nckap5l
|
NCK-associated protein 5-like |
| chr8_-_116965396 | 3.98 |
ENSRNOT00000042528
|
Bsn
|
bassoon (presynaptic cytomatrix protein) |
| chr5_+_166533181 | 3.89 |
ENSRNOT00000045063
|
Clstn1
|
calsyntenin 1 |
| chr10_-_63952726 | 3.89 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr11_+_86512797 | 3.81 |
ENSRNOT00000051680
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr1_+_163663407 | 3.73 |
ENSRNOT00000088873
|
Thap12
|
THAP domain containing 12 |
| chr14_+_75852060 | 3.67 |
ENSRNOT00000075975
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr17_-_80320681 | 3.64 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr10_-_89699836 | 3.42 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr5_-_64850427 | 3.41 |
ENSRNOT00000008883
|
Tmem246
|
transmembrane protein 246 |
| chr10_+_82800704 | 3.35 |
ENSRNOT00000089497
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr11_+_86421106 | 3.30 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr10_-_90393317 | 3.29 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr1_-_267245636 | 3.25 |
ENSRNOT00000082799
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr1_-_265798167 | 3.20 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr10_+_4957326 | 3.16 |
ENSRNOT00000003458
|
Socs1
|
suppressor of cytokine signaling 1 |
| chr12_+_47103314 | 2.93 |
ENSRNOT00000064910
|
Rnf10
|
ring finger protein 10 |
| chr11_+_87722350 | 2.91 |
ENSRNOT00000000313
|
Scarf2
|
scavenger receptor class F, member 2 |
| chr6_+_137997335 | 2.82 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr7_-_132757558 | 2.57 |
ENSRNOT00000021153
|
Slc2a13
|
solute carrier family 2 member 13 |
| chrX_+_120859968 | 2.55 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr4_-_163119391 | 2.54 |
ENSRNOT00000086605
|
Chtopl1
|
chromatin target of PRMT1-like 1 |
| chrX_+_15679254 | 2.48 |
ENSRNOT00000045256
|
Plp2
|
proteolipid protein 2 |
| chr6_-_109162267 | 2.47 |
ENSRNOT00000077518
|
Nek9
|
NIMA-related kinase 9 |
| chr7_+_130498199 | 2.47 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr7_+_53275676 | 2.40 |
ENSRNOT00000031986
|
E2f7
|
E2F transcription factor 7 |
| chr1_-_215836641 | 2.39 |
ENSRNOT00000080246
|
Igf2
|
insulin-like growth factor 2 |
| chr14_-_59323947 | 2.35 |
ENSRNOT00000004006
|
Stim2
|
stromal interaction molecule 2 |
| chr7_-_58286770 | 2.34 |
ENSRNOT00000005258
|
Rab21
|
RAB21, member RAS oncogene family |
| chrX_+_120860178 | 2.25 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr5_+_156876706 | 2.13 |
ENSRNOT00000021864
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr7_+_25919867 | 2.12 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr1_+_274766283 | 2.11 |
ENSRNOT00000071541
|
Adra2a
|
adrenoceptor alpha 2A |
| chr5_+_58393233 | 2.11 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr16_+_19051965 | 2.09 |
ENSRNOT00000016399
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr1_+_80087684 | 2.06 |
ENSRNOT00000041215
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr2_+_218951451 | 2.03 |
ENSRNOT00000019190
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr5_+_173183990 | 2.03 |
ENSRNOT00000068062
|
Tmem240
|
transmembrane protein 240 |
| chr17_+_9679628 | 2.03 |
ENSRNOT00000019569
ENSRNOT00000019393 |
Dbn1
|
drebrin 1 |
| chr3_+_4083864 | 2.02 |
ENSRNOT00000006186
|
Fam69b
|
family with sequence similarity 69, member B |
| chr10_+_75365822 | 1.98 |
ENSRNOT00000055705
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr8_-_49025917 | 1.95 |
ENSRNOT00000078816
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr18_-_63394690 | 1.94 |
ENSRNOT00000090078
ENSRNOT00000029431 |
Cep76
|
centrosomal protein 76 |
| chr8_-_68966108 | 1.93 |
ENSRNOT00000012155
|
Smad6
|
SMAD family member 6 |
| chr10_+_56591292 | 1.92 |
ENSRNOT00000023379
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr9_+_98924134 | 1.85 |
ENSRNOT00000027597
|
Twist2
|
twist family bHLH transcription factor 2 |
| chr7_+_141355994 | 1.83 |
ENSRNOT00000081195
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr19_+_18079094 | 1.83 |
ENSRNOT00000029415
|
Tox3
|
TOX high mobility group box family member 3 |
| chr8_+_48805684 | 1.82 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr14_-_37871051 | 1.81 |
ENSRNOT00000003087
|
Slain2
|
SLAIN motif family, member 2 |
| chr7_-_12085226 | 1.77 |
ENSRNOT00000088669
ENSRNOT00000079925 |
Onecut3
|
one cut domain, family member 3 |
| chr8_+_122197027 | 1.75 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr1_+_221612584 | 1.74 |
ENSRNOT00000090100
|
Atg2a
|
autophagy related 2A |
| chr1_-_254671596 | 1.70 |
ENSRNOT00000025450
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr2_+_218951141 | 1.69 |
ENSRNOT00000091001
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr8_-_116391158 | 1.59 |
ENSRNOT00000078720
ENSRNOT00000022550 |
Gnai2
|
G protein subunit alpha i2 |
| chr1_-_221486633 | 1.56 |
ENSRNOT00000028509
|
Sac3d1
|
SAC3 domain containing 1 |
| chr1_+_239398043 | 1.56 |
ENSRNOT00000087386
|
Tmem2
|
transmembrane protein 2 |
| chr7_+_120125633 | 1.56 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr3_+_114747410 | 1.53 |
ENSRNOT00000043414
|
Spata5l1
|
spermatogenesis associated 5-like 1 |
| chr5_+_58393603 | 1.52 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr6_-_46631983 | 1.48 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr3_+_146980923 | 1.46 |
ENSRNOT00000011654
|
Nsfl1c
|
NSFL1 cofactor |
| chr1_-_167347490 | 1.43 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr1_-_221015929 | 1.43 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr5_+_134593517 | 1.43 |
ENSRNOT00000013471
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr3_-_164095878 | 1.42 |
ENSRNOT00000079414
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chr2_-_206274079 | 1.41 |
ENSRNOT00000056079
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr4_+_157453069 | 1.39 |
ENSRNOT00000088622
|
Mlf2
|
myeloid leukemia factor 2 |
| chr3_-_1924827 | 1.36 |
ENSRNOT00000006162
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chr15_+_108908607 | 1.35 |
ENSRNOT00000089455
|
Zic2
|
Zic family member 2 |
| chr2_-_189899103 | 1.34 |
ENSRNOT00000010455
|
Chtop
|
chromatin target of PRMT1 |
| chr3_+_111049118 | 1.29 |
ENSRNOT00000088870
|
Spint1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr7_+_139271698 | 1.14 |
ENSRNOT00000079388
|
Slc48a1
|
solute carrier family 48 member 1 |
| chr9_-_44786041 | 1.14 |
ENSRNOT00000067197
|
Rev1
|
REV1, DNA directed polymerase |
| chr3_+_111049315 | 1.11 |
ENSRNOT00000017223
|
Spint1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr2_-_189899325 | 1.09 |
ENSRNOT00000017561
|
Chtop
|
chromatin target of PRMT1 |
| chr19_-_56054142 | 1.08 |
ENSRNOT00000051945
|
Vps9d1
|
VPS9 domain containing 1 |
| chr10_-_84698886 | 1.05 |
ENSRNOT00000067542
|
Nfe2l1
|
nuclear factor, erythroid 2-like 1 |
| chr1_+_228395558 | 0.97 |
ENSRNOT00000065411
|
Osbp
|
oxysterol binding protein |
| chr3_-_83306781 | 0.92 |
ENSRNOT00000014088
|
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr6_+_2569013 | 0.92 |
ENSRNOT00000008988
|
Atl2
|
atlastin GTPase 2 |
| chr5_+_150459713 | 0.92 |
ENSRNOT00000081681
ENSRNOT00000074251 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chrX_+_136466779 | 0.90 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chr10_-_90127600 | 0.89 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr12_-_46718355 | 0.87 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr4_+_157452607 | 0.84 |
ENSRNOT00000022467
|
Mlf2
|
myeloid leukemia factor 2 |
| chr4_-_115157263 | 0.83 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr1_-_254671778 | 0.81 |
ENSRNOT00000025493
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr7_-_63045728 | 0.74 |
ENSRNOT00000039532
|
Lemd3
|
LEM domain containing 3 |
| chr3_+_108795235 | 0.70 |
ENSRNOT00000007028
|
Spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr7_-_134560713 | 0.68 |
ENSRNOT00000006621
|
Yaf2
|
YY1 associated factor 2 |
| chr2_-_197935567 | 0.67 |
ENSRNOT00000085404
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr11_+_82466071 | 0.63 |
ENSRNOT00000036958
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_+_52086325 | 0.62 |
ENSRNOT00000020341
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr9_+_42620006 | 0.62 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr8_+_115069095 | 0.61 |
ENSRNOT00000014770
|
Dusp7
|
dual specificity phosphatase 7 |
| chr1_+_165724451 | 0.48 |
ENSRNOT00000025827
|
Fam168a
|
family with sequence similarity 168, member A |
| chr7_+_116671948 | 0.43 |
ENSRNOT00000077773
ENSRNOT00000029711 |
Gli4
|
GLI family zinc finger 4 |
| chr5_+_136683592 | 0.43 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr10_+_110469290 | 0.41 |
ENSRNOT00000054919
|
Foxk2
|
forkhead box K2 |
| chr15_-_25933268 | 0.41 |
ENSRNOT00000067735
|
Exoc5
|
exocyst complex component 5 |
| chr6_-_108415093 | 0.39 |
ENSRNOT00000031650
|
Syndig1l
|
synapse differentiation inducing 1-like |
| chr5_+_60250546 | 0.38 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr19_+_56179111 | 0.36 |
ENSRNOT00000046155
|
Tcf25
|
transcription factor 25 |
| chr2_-_24923128 | 0.34 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr17_+_72240802 | 0.33 |
ENSRNOT00000025993
ENSRNOT00000090453 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr17_-_32953641 | 0.30 |
ENSRNOT00000023332
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr10_-_82197848 | 0.28 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr17_-_10208360 | 0.27 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr10_-_35800120 | 0.27 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr1_-_164977633 | 0.26 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr6_-_22281886 | 0.25 |
ENSRNOT00000039375
|
Spast
|
spastin |
| chr19_+_56054212 | 0.24 |
ENSRNOT00000022324
|
Zfp276
|
zinc finger protein (C2H2 type) 276 |
| chr7_-_64341207 | 0.24 |
ENSRNOT00000006051
|
Tmem5
|
transmembrane protein 5 |
| chr6_+_136041777 | 0.21 |
ENSRNOT00000014395
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr13_+_90532326 | 0.19 |
ENSRNOT00000008944
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr10_+_29289203 | 0.19 |
ENSRNOT00000067013
|
Pwwp2a
|
PWWP domain containing 2A |
| chr5_-_152198813 | 0.16 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr18_+_86071662 | 0.12 |
ENSRNOT00000064901
|
Rttn
|
rotatin |
| chr1_-_222590112 | 0.11 |
ENSRNOT00000028763
ENSRNOT00000091990 |
Mark2
|
microtubule affinity regulating kinase 2 |
| chr2_-_210088949 | 0.11 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr1_-_127599257 | 0.10 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr8_+_117486083 | 0.09 |
ENSRNOT00000027552
ENSRNOT00000088705 |
Prkar2a
|
protein kinase cAMP-dependent type 2 regulatory subunit alpha |
| chr15_+_25933483 | 0.08 |
ENSRNOT00000018972
|
Ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr5_-_166726794 | 0.08 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr7_-_130408187 | 0.07 |
ENSRNOT00000015374
|
Chkb
|
choline kinase beta |
| chrX_+_69580015 | 0.07 |
ENSRNOT00000057920
|
Fam155b
|
family with sequence similarity 155, member B |
| chr18_-_28495937 | 0.05 |
ENSRNOT00000027194
|
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr6_+_136279496 | 0.04 |
ENSRNOT00000091946
|
Apopt1
|
apoptogenic 1, mitochondrial |
| chr2_+_198231291 | 0.04 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr15_-_4022223 | 0.01 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mbd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 1.6 | 6.3 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 1.4 | 5.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.3 | 1.3 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 1.2 | 3.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.1 | 3.3 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.0 | 7.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.9 | 5.6 | GO:0071306 | cellular response to vitamin E(GO:0071306) |
| 0.8 | 3.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.7 | 15.5 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.7 | 4.8 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.7 | 2.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.7 | 4.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.6 | 2.5 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.6 | 2.9 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.6 | 7.0 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.5 | 3.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.5 | 2.1 | GO:0071883 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) thermoception(GO:0050955) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.5 | 6.0 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.5 | 3.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.5 | 1.9 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.5 | 2.4 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.5 | 1.4 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.4 | 3.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.4 | 3.2 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.3 | 2.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 2.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.3 | 4.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.3 | 3.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 4.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 2.4 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.2 | 2.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.2 | 1.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 2.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.2 | 0.8 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.2 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 18.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.9 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 2.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.7 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 1.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.9 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.3 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 1.9 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 2.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.4 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.1 | 4.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.3 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.7 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.8 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) |
| 0.1 | 0.9 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 5.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 5.0 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 2.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 5.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 4.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 2.5 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 3.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 1.0 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 1.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 2.0 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 2.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 3.8 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 5.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 2.1 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 3.0 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 2.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.8 | 2.3 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.6 | 3.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.5 | 1.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.5 | 5.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 2.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.3 | 3.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 6.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 5.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 4.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 7.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 3.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.2 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 4.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 3.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 5.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.9 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) |
| 0.0 | 4.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 13.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 3.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 6.9 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 3.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 1.2 | 3.7 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 1.2 | 5.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.1 | 5.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.9 | 5.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 2.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.6 | 1.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.6 | 3.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 4.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.6 | 4.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.6 | 3.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.5 | 2.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 3.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 6.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 3.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 5.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 2.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 2.6 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 2.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 2.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 4.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 7.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 7.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.6 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 13.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 3.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 3.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 3.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 12.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0015187 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 3.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 2.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 5.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 7.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.4 | 8.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.4 | 10.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 3.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 2.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 4.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 6.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.5 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |