Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
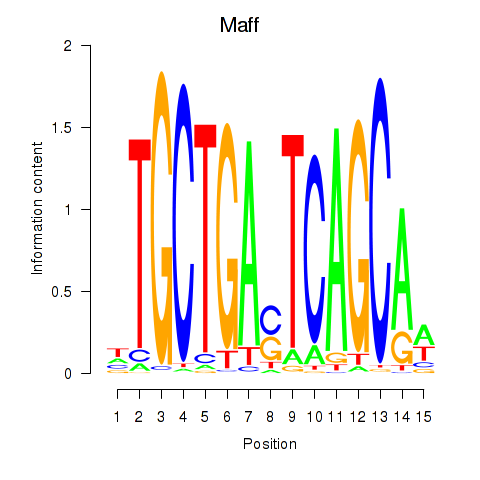
Results for Maff
Z-value: 0.60
Transcription factors associated with Maff
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maff
|
ENSRNOG00000012886 | MAF bZIP transcription factor F |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maff | rn6_v1_chr7_+_120580743_120580743 | -0.28 | 5.0e-07 | Click! |
Activity profile of Maff motif
Sorted Z-values of Maff motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_29171783 | 24.55 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr15_+_2527343 | 17.73 |
ENSRNOT00000089013
|
Dusp13
|
dual specificity phosphatase 13 |
| chr15_-_28104206 | 17.23 |
ENSRNOT00000032536
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr15_+_2734068 | 16.79 |
ENSRNOT00000017663
|
Dusp13
|
dual specificity phosphatase 13 |
| chr10_-_93675991 | 16.59 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr3_-_72289310 | 16.46 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr10_+_53818818 | 16.23 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr9_-_95290931 | 15.64 |
ENSRNOT00000025300
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr16_-_68832862 | 15.29 |
ENSRNOT00000016348
|
Poteg
|
POTE ankyrin domain family, member G |
| chr15_+_28028521 | 15.06 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr18_-_28454756 | 14.29 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr8_-_73164620 | 13.64 |
ENSRNOT00000031988
|
Tln2
|
talin 2 |
| chr7_+_51794173 | 13.39 |
ENSRNOT00000043774
|
Otogl
|
otogelin-like |
| chr7_-_124999137 | 13.11 |
ENSRNOT00000039228
|
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr14_+_34389991 | 12.42 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr5_-_19368431 | 10.33 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr16_-_20800646 | 10.17 |
ENSRNOT00000083281
|
Comp
|
cartilage oligomeric matrix protein |
| chr6_+_52702544 | 9.57 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr18_+_74006046 | 9.40 |
ENSRNOT00000086400
|
AABR07032622.1
|
|
| chr14_-_72380330 | 9.04 |
ENSRNOT00000082653
ENSRNOT00000081878 ENSRNOT00000006727 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr1_-_102013243 | 8.98 |
ENSRNOT00000042115
|
Abcc6
|
ATP binding cassette subfamily C member 6 |
| chr6_-_26281300 | 8.64 |
ENSRNOT00000078750
|
RGD1560110
|
similar to RIKEN cDNA 4930548H24 |
| chr18_+_40883224 | 7.87 |
ENSRNOT00000039733
|
Lvrn
|
laeverin |
| chr2_-_192960294 | 7.79 |
ENSRNOT00000012420
|
LOC102552326
|
late cornified envelope protein 5A-like |
| chr5_+_28850950 | 7.58 |
ENSRNOT00000009759
|
Tmem64
|
transmembrane protein 64 |
| chr6_+_54488038 | 7.37 |
ENSRNOT00000084661
ENSRNOT00000005637 |
Snx13
|
sorting nexin 13 |
| chrX_+_13441558 | 7.26 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr11_-_32469566 | 7.20 |
ENSRNOT00000002718
|
LOC498063
|
similar to RIKEN cDNA 4930563D23 |
| chr10_-_13168217 | 7.14 |
ENSRNOT00000087768
|
Elob
|
elongin B |
| chr9_+_8349033 | 6.75 |
ENSRNOT00000073775
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chr2_-_210943620 | 6.64 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr13_-_111765944 | 6.39 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr7_-_117267402 | 5.92 |
ENSRNOT00000088945
|
Plec
|
plectin |
| chr20_+_6351458 | 5.67 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chrX_-_123788898 | 5.60 |
ENSRNOT00000009123
|
Akap14
|
A-kinase anchoring protein 14 |
| chr9_+_47386626 | 5.47 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr16_-_14382641 | 5.30 |
ENSRNOT00000018723
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr8_+_128829680 | 5.25 |
ENSRNOT00000025279
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr6_-_86822094 | 4.97 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr7_-_117267803 | 4.53 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr14_+_42402081 | 4.17 |
ENSRNOT00000092933
|
Bend4
|
BEN domain containing 4 |
| chr3_-_147188929 | 4.11 |
ENSRNOT00000055455
|
Rad21l1
|
RAD21 cohesin complex component like 1 |
| chr9_-_82154266 | 4.05 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr5_+_127571312 | 3.63 |
ENSRNOT00000090817
ENSRNOT00000077808 ENSRNOT00000072263 |
Slc1a7
|
solute carrier family 1 member 7 |
| chr5_+_127571114 | 3.48 |
ENSRNOT00000016557
|
Slc1a7
|
solute carrier family 1 member 7 |
| chr5_-_28737719 | 3.26 |
ENSRNOT00000009718
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr8_-_1450138 | 3.25 |
ENSRNOT00000008062
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr10_+_87819150 | 3.25 |
ENSRNOT00000073764
ENSRNOT00000064626 |
LOC680442
|
hypothetical protein LOC680442 |
| chr8_-_109576353 | 3.08 |
ENSRNOT00000010320
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr1_-_87825333 | 3.01 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr4_-_114768029 | 2.99 |
ENSRNOT00000083404
|
Tlx2
|
T-cell leukemia homeobox 2 |
| chr4_-_113902399 | 2.94 |
ENSRNOT00000087788
|
Tlx2
|
T-cell leukemia homeobox 2 |
| chr2_-_193007929 | 2.78 |
ENSRNOT00000056486
|
LOC102552128
|
late cornified envelope protein 1C-like |
| chr6_-_75470347 | 2.75 |
ENSRNOT00000047882
|
RGD1559724
|
similar to 40S ribosomal protein S19 |
| chr3_-_11410732 | 2.69 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr2_+_208373154 | 2.66 |
ENSRNOT00000082943
ENSRNOT00000050538 |
LOC100911796
|
adenosine receptor A3-like |
| chr2_-_14701903 | 2.66 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr2_-_153140894 | 2.62 |
ENSRNOT00000075166
|
AABR07010840.1
|
|
| chr1_-_88240057 | 2.61 |
ENSRNOT00000073092
|
LOC103690038
|
zinc finger protein 569 |
| chr2_-_193042323 | 2.58 |
ENSRNOT00000012464
|
AABR07012317.1
|
|
| chr15_-_109394905 | 2.57 |
ENSRNOT00000019602
ENSRNOT00000078424 |
Tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr19_-_43848937 | 2.52 |
ENSRNOT00000044844
|
Ldhd
|
lactate dehydrogenase D |
| chr2_+_160291450 | 2.48 |
ENSRNOT00000075563
|
AABR07011278.1
|
|
| chrX_-_107762590 | 2.45 |
ENSRNOT00000072739
|
Esx1
|
ESX homeobox 1 |
| chr15_-_28786094 | 2.45 |
ENSRNOT00000044250
|
Olr1639
|
olfactory receptor 1639 |
| chr8_-_3882077 | 2.22 |
ENSRNOT00000071085
|
Vom2r25
|
vomeronasal 2 receptor, 25 |
| chr10_+_34712870 | 2.22 |
ENSRNOT00000045600
|
Olr1396
|
olfactory receptor 1396 |
| chrX_+_15321713 | 2.19 |
ENSRNOT00000071835
|
LOC108348138
|
GTPase ERas |
| chr10_+_12602388 | 2.14 |
ENSRNOT00000060989
|
Olr1373
|
olfactory receptor 1373 |
| chr8_+_408001 | 2.05 |
ENSRNOT00000046058
|
Gucy1a2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr15_+_33596301 | 1.96 |
ENSRNOT00000022563
|
Il25
|
interleukin 25 |
| chr17_-_38858331 | 1.91 |
ENSRNOT00000059738
|
Prl2c1
|
Prolactin family 2, subfamily c, member 1 |
| chr1_+_228337767 | 1.88 |
ENSRNOT00000066247
|
Patl1
|
PAT1 homolog 1, processing body mRNA decay factor |
| chr15_-_28806752 | 1.85 |
ENSRNOT00000047436
|
Olr1640
|
olfactory receptor 1640 |
| chr13_-_79899479 | 1.85 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chr20_-_11634137 | 1.64 |
ENSRNOT00000083064
|
LOC100361793
|
keratin associated protein 12-1-like |
| chr12_-_50404550 | 1.33 |
ENSRNOT00000073072
|
Crybb1
|
crystallin, beta B1 |
| chr10_-_74001895 | 1.27 |
ENSRNOT00000005658
|
Vmp1
|
vacuole membrane protein 1 |
| chr14_-_21127868 | 1.23 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr11_+_71151132 | 1.06 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr1_+_31700953 | 1.05 |
ENSRNOT00000020251
|
Exoc3
|
exocyst complex component 3 |
| chr10_+_87808493 | 0.98 |
ENSRNOT00000065405
|
LOC680428
|
hypothetical protein LOC680428 |
| chr4_+_147832136 | 0.96 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr4_-_117568348 | 0.95 |
ENSRNOT00000071447
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr2_-_193101051 | 0.93 |
ENSRNOT00000065242
|
AABR07012318.1
|
|
| chr1_-_170073473 | 0.88 |
ENSRNOT00000085141
|
Olr196
|
olfactory receptor 196 |
| chrX_-_45806198 | 0.84 |
ENSRNOT00000066663
|
Prrg1
|
proline rich and Gla domain 1 |
| chr1_-_221448570 | 0.84 |
ENSRNOT00000075165
|
Zfpl1
|
zinc finger protein-like 1 |
| chr5_+_48224994 | 0.71 |
ENSRNOT00000068028
|
Rragd
|
Ras-related GTP binding D |
| chrX_-_10772633 | 0.70 |
ENSRNOT00000035153
|
Ube2q2l
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr1_-_88826302 | 0.61 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr6_-_49089855 | 0.59 |
ENSRNOT00000006526
|
Tpo
|
thyroid peroxidase |
| chr13_-_27158628 | 0.57 |
ENSRNOT00000003475
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr13_-_36101411 | 0.57 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr6_-_78817641 | 0.54 |
ENSRNOT00000072117
|
AABR07064392.1
|
|
| chr19_-_49510901 | 0.54 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr1_-_89483988 | 0.53 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr16_+_19881122 | 0.53 |
ENSRNOT00000060404
|
Dda1
|
DET1 and DDB1 associated 1 |
| chr1_+_87009730 | 0.49 |
ENSRNOT00000027537
|
Ech1
|
enoyl-CoA hydratase 1 |
| chr10_-_43931094 | 0.49 |
ENSRNOT00000039778
|
Olr1415
|
olfactory receptor 1415 |
| chr1_-_99985422 | 0.30 |
ENSRNOT00000025701
|
Klk1c2
|
kallikrein 1-related peptidase C2 |
| chr10_+_60043848 | 0.27 |
ENSRNOT00000080593
|
AABR07029918.1
|
|
| chr7_-_3499697 | 0.17 |
ENSRNOT00000064311
|
Olr878
|
olfactory receptor 878 |
| chr7_+_16394354 | 0.13 |
ENSRNOT00000041157
|
Olr1058
|
olfactory receptor 1058 |
| chr10_+_87832743 | 0.04 |
ENSRNOT00000055286
|
Krtap31-1
|
keratin associated protein 31-1 |
| chr7_+_91384187 | 0.01 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
Network of associatons between targets according to the STRING database.
First level regulatory network of Maff
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 24.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 2.3 | 9.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 1.9 | 7.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 1.6 | 12.4 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 1.5 | 16.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 1.2 | 13.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.2 | 13.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 1.0 | 3.1 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.8 | 5.7 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.8 | 10.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.7 | 16.5 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.7 | 3.3 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.6 | 7.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 10.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.5 | 10.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.4 | 5.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 7.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.4 | 4.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 1.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.3 | 5.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.3 | 13.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 2.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.3 | 34.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 10.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 7.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 2.7 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 1.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 2.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 1.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.7 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 17.2 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 6.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 5.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 4.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 15.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 18.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 40.8 | GO:0032982 | myosin filament(GO:0032982) |
| 1.9 | 5.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.0 | 7.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.7 | 13.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.6 | 4.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 10.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 9.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 10.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 16.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 5.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 11.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 8.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 15.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 3.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 2.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 5.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.3 | GO:0000421 | pre-autophagosomal structure(GO:0000407) autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 8.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 6.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 5.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 6.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.1 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.9 | 24.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.7 | 10.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.6 | 12.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.7 | 16.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.7 | 9.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.7 | 13.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.6 | 34.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.5 | 5.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 7.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.4 | 5.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 5.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 10.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.4 | 7.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 7.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 10.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 7.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 16.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 17.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 3.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 13.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 13.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 1.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 5.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 6.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 16.6 | GO:0016874 | ligase activity(GO:0016874) |
| 0.1 | 2.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 9.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 7.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 15.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.2 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 5.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 7.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 14.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 5.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 6.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 40.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.7 | 10.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.4 | 10.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 5.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 7.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 3.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 9.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |