Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
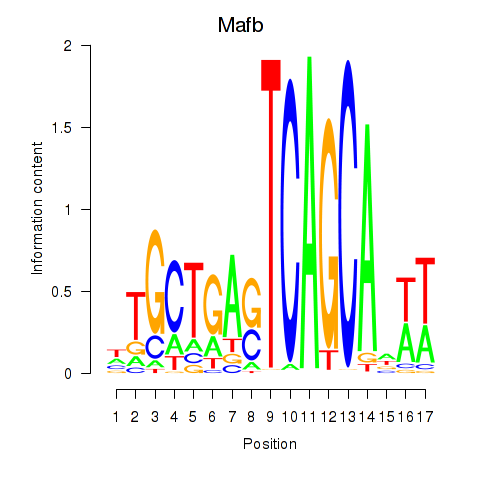
Results for Mafb
Z-value: 1.01
Transcription factors associated with Mafb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mafb
|
ENSRNOG00000016037 | MAF bZIP transcription factor B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafb | rn6_v1_chr3_-_156340913_156340913 | 0.01 | 8.3e-01 | Click! |
Activity profile of Mafb motif
Sorted Z-values of Mafb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_38828444 | 36.02 |
ENSRNOT00000001809
|
Hpd
|
4-hydroxyphenylpyruvate dioxygenase |
| chr17_+_21382455 | 32.37 |
ENSRNOT00000019756
|
Elovl2
|
ELOVL fatty acid elongase 2 |
| chr11_-_81444375 | 30.63 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr13_+_89597138 | 30.34 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr7_+_28066635 | 28.92 |
ENSRNOT00000005844
|
Pah
|
phenylalanine hydroxylase |
| chr14_-_19072677 | 25.35 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr1_-_240601744 | 23.57 |
ENSRNOT00000024093
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr1_-_224698514 | 23.55 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr14_+_22553650 | 22.21 |
ENSRNOT00000092201
ENSRNOT00000002712 |
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr1_+_238222521 | 21.65 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr7_+_116739062 | 21.09 |
ENSRNOT00000075837
ENSRNOT00000076670 ENSRNOT00000010065 |
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr1_-_162713610 | 19.76 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr16_+_6078122 | 19.33 |
ENSRNOT00000021407
|
Chdh
|
choline dehydrogenase |
| chr4_+_154215250 | 17.90 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr10_-_38774449 | 17.63 |
ENSRNOT00000049820
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr7_+_11490852 | 16.98 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr15_-_28104206 | 16.71 |
ENSRNOT00000032536
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr8_+_48718329 | 16.68 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr15_+_4077091 | 16.35 |
ENSRNOT00000011554
|
Myoz1
|
myozenin 1 |
| chr15_+_4077951 | 16.25 |
ENSRNOT00000085266
|
Myoz1
|
myozenin 1 |
| chr5_-_9429859 | 16.12 |
ENSRNOT00000009462
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr18_-_15089988 | 15.97 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr13_-_36101411 | 15.91 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr11_+_36743901 | 15.54 |
ENSRNOT00000085014
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr10_+_53781239 | 15.50 |
ENSRNOT00000082871
|
Myh2
|
myosin heavy chain 2 |
| chr10_+_56662242 | 15.29 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr5_-_19368431 | 15.29 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr4_-_117568348 | 15.26 |
ENSRNOT00000071447
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr1_+_106896790 | 15.04 |
ENSRNOT00000042952
ENSRNOT00000064193 |
Ano5
|
anoctamin 5 |
| chr9_+_49479023 | 14.93 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr10_+_56662561 | 14.75 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr10_-_7200499 | 14.74 |
ENSRNOT00000003633
|
Abat
|
4-aminobutyrate aminotransferase |
| chr19_-_25134055 | 14.55 |
ENSRNOT00000007519
|
Misp3
|
MISP family member 3 |
| chr16_-_49003246 | 14.46 |
ENSRNOT00000089501
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr11_+_61609370 | 14.10 |
ENSRNOT00000088880
ENSRNOT00000082533 |
Gramd1c
|
GRAM domain containing 1C |
| chr7_+_101069104 | 14.06 |
ENSRNOT00000056894
|
AABR07058124.2
|
|
| chr13_-_56877611 | 13.96 |
ENSRNOT00000079040
ENSRNOT00000017195 |
Cfhr1
|
complement factor H-related 1 |
| chr4_-_30338679 | 13.90 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr1_-_219312240 | 13.90 |
ENSRNOT00000066691
|
RGD1307603
|
similar to hypothetical protein MGC37914 |
| chr5_+_160306727 | 13.73 |
ENSRNOT00000016648
|
Agmat
|
agmatinase |
| chr11_-_87921679 | 13.66 |
ENSRNOT00000084973
|
Serpind1
|
serpin family D member 1 |
| chr12_-_19167015 | 13.43 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr15_-_3650819 | 13.15 |
ENSRNOT00000014273
|
Plau
|
plasminogen activator, urokinase |
| chr2_+_127845034 | 13.06 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr6_+_8284878 | 12.99 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr6_-_129010271 | 12.31 |
ENSRNOT00000075378
|
Serpina10
|
serpin family A member 10 |
| chr18_-_55859333 | 12.15 |
ENSRNOT00000025989
|
Myoz3
|
myozenin 3 |
| chr11_-_87924816 | 11.93 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr7_-_71226150 | 11.91 |
ENSRNOT00000005875
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr9_-_73948583 | 11.82 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr1_-_99303048 | 11.34 |
ENSRNOT00000025631
|
LOC687880
|
similar to Kallikrein-1 precursor (Tissue kallikrein) (Kidney/pancreas/salivary gland kallikrein) |
| chr1_-_221919210 | 11.28 |
ENSRNOT00000092580
ENSRNOT00000028665 |
Slc22a12
|
solute carrier family 22 member 12 |
| chr14_+_2050483 | 11.26 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chr1_-_99870024 | 11.21 |
ENSRNOT00000085556
|
AABR07003241.2
|
|
| chr11_+_44089797 | 11.16 |
ENSRNOT00000060861
|
LOC108352322
|
ferritin heavy chain-like |
| chr13_+_75177965 | 11.07 |
ENSRNOT00000007321
|
Sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr2_+_243502073 | 11.06 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_+_32199810 | 10.99 |
ENSRNOT00000036714
|
Slc6a19
|
solute carrier family 6 member 19 |
| chr2_+_78103387 | 10.93 |
ENSRNOT00000063951
|
LOC103689968
|
protein FAM134B |
| chr1_+_264507985 | 10.91 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chr13_-_56693968 | 10.82 |
ENSRNOT00000060160
|
AABR07021096.1
|
|
| chr10_+_53818818 | 10.71 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr1_+_221236773 | 10.43 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr5_+_152559577 | 10.40 |
ENSRNOT00000082245
|
Slc30a2
|
solute carrier family 30 member 2 |
| chr6_+_107517668 | 10.23 |
ENSRNOT00000013753
|
Acot4
|
acyl-CoA thioesterase 4 |
| chr5_+_137189473 | 10.13 |
ENSRNOT00000056815
|
Hyi
|
hydroxypyruvate isomerase |
| chrX_-_110230610 | 10.08 |
ENSRNOT00000093401
|
Serpina7
|
serpin family A member 7 |
| chr8_-_119012671 | 9.87 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr6_+_127766470 | 9.75 |
ENSRNOT00000013138
|
Serpina5
|
serpin family A member 5 |
| chr2_+_251634431 | 9.74 |
ENSRNOT00000045016
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr1_-_219311502 | 9.73 |
ENSRNOT00000085439
|
RGD1307603
|
similar to hypothetical protein MGC37914 |
| chrX_-_139079336 | 9.52 |
ENSRNOT00000047605
|
Usp26
|
ubiquitin specific peptidase 26 |
| chr12_+_16913312 | 9.39 |
ENSRNOT00000001718
|
Tmem184a
|
transmembrane protein 184A |
| chr7_-_124999137 | 9.39 |
ENSRNOT00000039228
|
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr2_-_84531192 | 9.18 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr2_-_195935878 | 9.18 |
ENSRNOT00000028440
|
Cgn
|
cingulin |
| chr1_-_275876329 | 9.16 |
ENSRNOT00000047903
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr2_-_93641497 | 9.14 |
ENSRNOT00000013720
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr3_+_56355431 | 9.11 |
ENSRNOT00000037188
|
Myo3b
|
myosin IIIB |
| chr10_-_34333305 | 9.08 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr3_+_143172686 | 9.06 |
ENSRNOT00000006869
|
Cst9l
|
cystatin 9-like |
| chr3_+_147511406 | 8.93 |
ENSRNOT00000082438
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr15_+_28028521 | 8.71 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr10_-_82229140 | 8.59 |
ENSRNOT00000004400
|
Epn3
|
epsin 3 |
| chr2_+_242882306 | 8.40 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr10_+_110346453 | 8.38 |
ENSRNOT00000054928
|
Tex19.1
|
testis expressed 19.1 |
| chr9_+_95233957 | 8.37 |
ENSRNOT00000071003
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr13_-_48927483 | 8.26 |
ENSRNOT00000010976
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr15_-_5509839 | 8.22 |
ENSRNOT00000080252
ENSRNOT00000081979 ENSRNOT00000085137 ENSRNOT00000049746 |
Spetex-2F
|
Spetex-2F protein |
| chr18_-_15225427 | 8.17 |
ENSRNOT00000090853
|
Rnf125
|
ring finger protein 125 |
| chr1_-_250727079 | 8.11 |
ENSRNOT00000079942
ENSRNOT00000087588 |
Sgms1
|
sphingomyelin synthase 1 |
| chr3_+_151032952 | 8.07 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr18_-_77579969 | 8.02 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr11_+_57404196 | 8.01 |
ENSRNOT00000042754
ENSRNOT00000034097 |
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr6_+_111180108 | 7.92 |
ENSRNOT00000082027
|
Gstz1
|
glutathione S-transferase zeta 1 |
| chr13_+_85580828 | 7.78 |
ENSRNOT00000005611
|
Aldh9a1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr4_+_22082194 | 7.72 |
ENSRNOT00000091799
|
Crot
|
carnitine O-octanoyltransferase |
| chrX_-_40086870 | 7.65 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr7_-_132143470 | 7.65 |
ENSRNOT00000038946
ENSRNOT00000044092 ENSRNOT00000066528 ENSRNOT00000045553 ENSRNOT00000046744 ENSRNOT00000055742 |
Kif21a
|
kinesin family member 21A |
| chr6_-_108120579 | 7.65 |
ENSRNOT00000041163
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr10_-_93675991 | 7.58 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr13_-_47377703 | 7.52 |
ENSRNOT00000005461
|
C4bpa
|
complement component 4 binding protein, alpha |
| chr8_-_116438038 | 7.52 |
ENSRNOT00000023791
|
Gnat1
|
G protein subunit alpha transducin 1 |
| chrX_+_13441558 | 7.51 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr6_+_36089433 | 7.46 |
ENSRNOT00000090438
ENSRNOT00000005731 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr10_-_46404642 | 7.43 |
ENSRNOT00000083698
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr1_+_277689729 | 7.32 |
ENSRNOT00000051834
|
Vwa2
|
von Willebrand factor A domain containing 2 |
| chr5_-_131860637 | 7.31 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr1_+_88955135 | 7.26 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr1_-_71710374 | 7.19 |
ENSRNOT00000078556
ENSRNOT00000046152 |
Nlrp4
|
NLR family, pyrin domain containing 4 |
| chr8_-_70932986 | 6.97 |
ENSRNOT00000042353
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr8_+_106449321 | 6.97 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr5_+_164845925 | 6.80 |
ENSRNOT00000011384
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr1_+_101397828 | 6.80 |
ENSRNOT00000028189
|
Kcna7
|
potassium voltage-gated channel subfamily A member 7 |
| chr20_+_4178686 | 6.73 |
ENSRNOT00000080641
|
Tesb
|
testis specific basic protein |
| chr10_+_34185898 | 6.60 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr2_+_22950018 | 6.51 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_150310319 | 6.47 |
ENSRNOT00000042081
|
Olr34
|
olfactory receptor 34 |
| chr2_-_188413219 | 6.40 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chr16_+_72086878 | 6.34 |
ENSRNOT00000023756
ENSRNOT00000078085 |
Adam3a
|
ADAM metallopeptidase domain 3A |
| chr10_-_56412544 | 6.30 |
ENSRNOT00000020578
|
Tmem102
|
transmembrane protein 102 |
| chr7_+_51794173 | 6.25 |
ENSRNOT00000043774
|
Otogl
|
otogelin-like |
| chr17_-_42127678 | 6.16 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_-_219259448 | 6.16 |
ENSRNOT00000024517
|
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr5_-_78042556 | 6.11 |
ENSRNOT00000076005
|
Rn50_5_0819.1
|
|
| chr8_+_49713190 | 6.09 |
ENSRNOT00000022074
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr11_-_32469566 | 6.05 |
ENSRNOT00000002718
|
LOC498063
|
similar to RIKEN cDNA 4930563D23 |
| chr8_-_40883880 | 5.96 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr2_-_88660449 | 5.96 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr9_+_99795678 | 5.87 |
ENSRNOT00000056601
|
Olr1353
|
olfactory receptor 1353 |
| chr4_+_180062799 | 5.86 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr6_-_60124274 | 5.85 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr2_-_192780631 | 5.85 |
ENSRNOT00000012371
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr1_-_213921208 | 5.83 |
ENSRNOT00000044393
|
Ano9
|
anoctamin 9 |
| chr6_-_39363367 | 5.75 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr10_+_4953879 | 5.69 |
ENSRNOT00000003455
|
Tnp2
|
transition protein 2 |
| chr16_+_19097391 | 5.69 |
ENSRNOT00000018018
|
Calr3
|
calreticulin 3 |
| chr20_-_33322966 | 5.68 |
ENSRNOT00000000459
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr5_-_171327450 | 5.68 |
ENSRNOT00000088710
|
Ccdc27
|
coiled-coil domain containing 27 |
| chr4_-_117758135 | 5.55 |
ENSRNOT00000087750
|
Nat8f4
|
N-acetyltransferase 8 (GCN5-related) family member 4 |
| chr4_+_120672152 | 5.54 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr5_-_77248563 | 5.53 |
ENSRNOT00000052243
|
LOC259245
|
alpha-2u globulin PGCL5 |
| chr1_-_143392532 | 5.41 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr16_-_6077978 | 5.40 |
ENSRNOT00000020823
|
Il17rb
|
interleukin 17 receptor B |
| chr3_-_146396299 | 5.38 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr1_-_167911961 | 5.36 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr19_-_20508711 | 5.34 |
ENSRNOT00000032408
|
LOC680913
|
hypothetical protein LOC680913 |
| chr1_-_99985422 | 5.13 |
ENSRNOT00000025701
|
Klk1c2
|
kallikrein 1-related peptidase C2 |
| chr1_+_99888297 | 5.07 |
ENSRNOT00000080896
|
AABR07003241.3
|
|
| chr4_-_117575154 | 5.02 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr5_+_155794229 | 5.00 |
ENSRNOT00000018127
|
LOC690206
|
hypothetical protein LOC690206 |
| chr4_-_117531480 | 4.98 |
ENSRNOT00000021120
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr7_-_18180705 | 4.94 |
ENSRNOT00000065954
|
Zfp494
|
zinc finger protein 494 |
| chr6_+_110749705 | 4.77 |
ENSRNOT00000084348
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr4_-_181348038 | 4.77 |
ENSRNOT00000082879
|
LOC690784
|
hypothetical protein LOC690783 |
| chr2_-_227207584 | 4.66 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr3_+_143151739 | 4.62 |
ENSRNOT00000006850
|
Cst13
|
cystatin 13 |
| chrM_+_7758 | 4.61 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr10_-_10807079 | 4.54 |
ENSRNOT00000087029
|
LOC360479
|
similar to hypothetical protein |
| chr15_-_12262164 | 4.48 |
ENSRNOT00000008798
|
Olr1606
|
olfactory receptor 1606 |
| chr8_+_84945444 | 4.48 |
ENSRNOT00000008244
|
Klhl31
|
kelch-like family member 31 |
| chr1_-_206394346 | 4.45 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr10_+_57064482 | 4.34 |
ENSRNOT00000046807
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr15_-_47442664 | 4.34 |
ENSRNOT00000072994
|
Prss55
|
protease, serine, 55 |
| chr19_+_683484 | 4.33 |
ENSRNOT00000090332
|
Terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr9_+_51009116 | 4.30 |
ENSRNOT00000039313
|
Mettl21cl1
|
methyltransferase like 21C-like 1 |
| chr5_+_126334803 | 4.30 |
ENSRNOT00000010288
|
Fam151a
|
family with sequence similarity 151, member A |
| chr17_+_16340226 | 4.28 |
ENSRNOT00000089140
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_42329232 | 4.28 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr2_+_154921999 | 4.27 |
ENSRNOT00000057620
|
LOC691044
|
similar to GTPase activating protein testicular GAP1 |
| chr9_+_24066303 | 4.23 |
ENSRNOT00000018163
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr10_-_90410569 | 4.19 |
ENSRNOT00000036112
|
Itga2b
|
integrin subunit alpha 2b |
| chr3_-_120011364 | 4.19 |
ENSRNOT00000018922
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr4_-_27698037 | 4.17 |
ENSRNOT00000038136
|
Pex1
|
peroxisomal biogenesis factor 1 |
| chrX_+_115721251 | 4.11 |
ENSRNOT00000060090
|
Trpc5os
|
TRPC5 opposite strand |
| chr10_+_88992487 | 4.10 |
ENSRNOT00000027061
|
Coasy
|
Coenzyme A synthase |
| chr15_+_28050975 | 4.07 |
ENSRNOT00000050944
|
LOC103693384
|
epididymal secretory protein E3-beta-like |
| chr2_-_260108897 | 4.07 |
ENSRNOT00000014106
|
Msh4
|
mutS homolog 4 |
| chr9_+_93445002 | 4.03 |
ENSRNOT00000025029
|
LOC501180
|
similar to hypothetical protein MGC35154 |
| chr10_+_63677396 | 4.00 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr1_-_12952906 | 3.96 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr1_-_82452281 | 3.96 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr4_+_52199416 | 3.92 |
ENSRNOT00000009537
|
Spam1
|
sperm adhesion molecule 1 |
| chrM_+_11736 | 3.91 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr14_+_77079402 | 3.91 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr6_+_52702544 | 3.86 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr13_+_88286718 | 3.85 |
ENSRNOT00000046076
|
LOC100366054
|
Da1-10-like |
| chr15_-_12129220 | 3.83 |
ENSRNOT00000029318
|
Olr1605
|
olfactory receptor 1605 |
| chr9_-_104467973 | 3.81 |
ENSRNOT00000026099
|
Slco6b1
|
solute carrier organic anion transporter family member 6B1 |
| chr1_+_83573032 | 3.77 |
ENSRNOT00000083650
|
AC142154.2
|
|
| chr8_-_72842228 | 3.76 |
ENSRNOT00000090288
|
Tpm1
|
tropomyosin 1, alpha |
| chr12_+_24761210 | 3.74 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr18_-_81682206 | 3.70 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr15_-_28092023 | 3.67 |
ENSRNOT00000090835
|
Eddm3b
|
epididymal protein 3B |
| chr3_+_15749691 | 3.67 |
ENSRNOT00000030984
|
Olr396
|
olfactory receptor 396 |
| chr10_-_87954055 | 3.62 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr8_-_132790778 | 3.58 |
ENSRNOT00000008255
ENSRNOT00000091431 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr14_+_60657686 | 3.55 |
ENSRNOT00000070892
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr3_-_52361060 | 3.54 |
ENSRNOT00000007832
|
Ttc21b
|
tetratricopeptide repeat domain 21B |
| chr8_-_90962470 | 3.50 |
ENSRNOT00000086426
|
Lca5
|
LCA5, lebercilin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mafb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.1 | 30.3 | GO:0046340 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 9.1 | 72.9 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 6.5 | 32.4 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 5.2 | 25.8 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 4.7 | 23.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 4.4 | 13.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 4.0 | 19.8 | GO:0015793 | glycerol transport(GO:0015793) |
| 3.6 | 21.6 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 3.4 | 10.2 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 3.4 | 27.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 3.2 | 34.8 | GO:0015747 | urate transport(GO:0015747) |
| 3.1 | 6.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 3.0 | 8.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 2.9 | 14.7 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.7 | 13.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 2.7 | 8.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 2.5 | 7.5 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 2.4 | 7.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 2.4 | 9.8 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.4 | 7.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.4 | 16.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 2.3 | 9.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 2.3 | 13.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.3 | 9.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 2.2 | 15.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 2.2 | 10.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 2.1 | 10.4 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.9 | 5.8 | GO:2001226 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 1.9 | 7.7 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 1.8 | 11.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.8 | 5.4 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 1.7 | 17.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.6 | 11.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.5 | 18.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 1.4 | 18.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.4 | 5.5 | GO:2000124 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.4 | 8.3 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 1.3 | 14.5 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 1.3 | 6.4 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 1.3 | 7.5 | GO:0050917 | sensory perception of umami taste(GO:0050917) positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 1.2 | 8.4 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 1.2 | 15.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.2 | 15.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 1.2 | 5.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.2 | 8.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 1.1 | 3.4 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 1.1 | 5.4 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 1.0 | 3.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.0 | 9.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 1.0 | 14.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.9 | 5.7 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.9 | 8.4 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.9 | 10.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.9 | 3.6 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.9 | 7.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.8 | 9.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.8 | 3.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.8 | 2.4 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.7 | 30.6 | GO:0042311 | vasodilation(GO:0042311) |
| 0.7 | 5.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.7 | 8.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.7 | 10.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.7 | 2.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.7 | 7.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.7 | 3.4 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 0.7 | 11.2 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.6 | 6.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.6 | 2.5 | GO:2000304 | cardiolipin acyl-chain remodeling(GO:0035965) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.6 | 6.8 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.6 | 1.2 | GO:1904404 | response to formaldehyde(GO:1904404) |
| 0.6 | 1.8 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.6 | 0.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.6 | 7.6 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.6 | 34.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.5 | 9.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.5 | 13.6 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.5 | 2.4 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.5 | 1.9 | GO:1900625 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) positive regulation of hyaluronan biosynthetic process(GO:1900127) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.5 | 14.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.4 | 2.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.4 | 8.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.4 | 3.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.4 | 6.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 2.1 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.4 | 2.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 2.4 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.4 | 1.9 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 8.0 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.4 | 12.6 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.4 | 1.5 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) |
| 0.4 | 6.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.4 | 2.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.4 | 3.5 | GO:0035721 | forebrain dorsal/ventral pattern formation(GO:0021798) intraciliary retrograde transport(GO:0035721) |
| 0.4 | 1.8 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.3 | 1.7 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.3 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 3.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 1.3 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.3 | 8.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 4.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.3 | 6.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.3 | 1.4 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.3 | 0.8 | GO:0044726 | protection of DNA demethylation of female pronucleus(GO:0044726) |
| 0.3 | 0.8 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.3 | 0.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 1.8 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.3 | 6.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 6.3 | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901028) |
| 0.2 | 4.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 4.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 8.2 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.2 | 0.7 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.2 | 0.4 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.2 | 8.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 9.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 10.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 1.9 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.2 | 52.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 0.4 | GO:2000017 | regulation of determination of dorsal identity(GO:2000015) positive regulation of determination of dorsal identity(GO:2000017) |
| 0.2 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 3.1 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.2 | 1.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 4.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 4.5 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 3.0 | GO:0021984 | spinal cord association neuron differentiation(GO:0021527) adenohypophysis development(GO:0021984) |
| 0.2 | 1.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 9.2 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.2 | 1.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 3.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 1.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 3.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 16.0 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.1 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.6 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 5.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.6 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.8 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 1.9 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 2.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 2.1 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.9 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 2.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 3.0 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 101.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 2.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 1.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 12.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 2.3 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.1 | 7.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.3 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 6.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 0.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.5 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.1 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.2 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.1 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 7.3 | GO:0007369 | gastrulation(GO:0007369) |
| 0.1 | 6.7 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 2.5 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 2.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 4.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 10.7 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.3 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 3.9 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 2.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 1.6 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 2.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.0 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 2.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 30.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.9 | 9.6 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.7 | 15.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.5 | 7.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.4 | 9.8 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 1.4 | 4.1 | GO:0005713 | recombination nodule(GO:0005713) MutSalpha complex(GO:0032301) |
| 1.3 | 9.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.9 | 8.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.9 | 8.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.8 | 4.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.7 | 9.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.6 | 13.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 2.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.6 | 52.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.6 | 8.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 3.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 3.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.5 | 6.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 3.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 1.5 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.4 | 4.3 | GO:0070187 | telosome(GO:0070187) |
| 0.4 | 6.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 8.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 18.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.4 | 4.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 10.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 38.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 2.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 44.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 5.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 18.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.3 | 27.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.3 | 1.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.3 | 4.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.0 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.2 | 0.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 29.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.2 | 11.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 8.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 116.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 2.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 9.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 2.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 3.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 49.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 8.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 3.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 3.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 5.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 20.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 2.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 6.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 6.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 97.5 | GO:1903561 | extracellular vesicle(GO:1903561) |
| 0.0 | 3.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 2.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 55.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.4 | 43.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 10.1 | 30.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 9.6 | 28.9 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 7.5 | 37.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 5.6 | 22.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 4.6 | 13.9 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 3.9 | 38.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 3.4 | 27.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 3.2 | 9.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 3.2 | 32.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 3.0 | 8.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 2.6 | 7.7 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 2.6 | 17.9 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 2.4 | 9.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 2.4 | 7.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.9 | 7.6 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 1.6 | 8.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.5 | 9.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.4 | 19.8 | GO:0015250 | water channel activity(GO:0015250) |
| 1.4 | 13.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.3 | 5.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.3 | 10.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 1.3 | 6.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.3 | 6.4 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.3 | 10.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.0 | 10.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 1.0 | 30.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.0 | 27.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 1.0 | 15.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.9 | 20.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.9 | 2.8 | GO:0031763 | galanin receptor binding(GO:0031763) type 2 galanin receptor binding(GO:0031765) type 3 galanin receptor binding(GO:0031766) |
| 0.9 | 18.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.9 | 32.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.8 | 2.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.8 | 4.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.8 | 17.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.8 | 2.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.8 | 7.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 7.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.8 | 6.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.8 | 3.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.7 | 11.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.7 | 7.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 5.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.7 | 7.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.7 | 4.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.7 | 3.4 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.7 | 49.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.6 | 14.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.6 | 7.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.6 | 5.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 14.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.5 | 15.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.5 | 2.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.5 | 3.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.5 | 14.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 2.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.5 | 54.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 2.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 3.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.5 | 9.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.5 | 6.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 1.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.5 | 10.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.5 | 1.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 1.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.4 | 3.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) transferrin receptor binding(GO:1990459) |
| 0.4 | 1.8 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.4 | 4.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.4 | 7.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 7.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 3.9 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.4 | 6.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.4 | 3.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 10.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 1.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 9.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 1.8 | GO:0016918 | retinal binding(GO:0016918) |
| 0.3 | 2.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 0.8 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.3 | 2.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.3 | 0.8 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 1.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.3 | 1.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 18.1 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.2 | 3.4 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.2 | 0.7 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.2 | 6.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 19.3 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.2 | 1.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 8.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 1.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 5.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 2.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 10.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 1.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 31.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 16.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 11.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 4.1 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 46.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 12.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 5.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 8.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 5.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 2.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 6.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.2 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.1 | 61.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 3.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 3.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 6.6 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.1 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 4.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 8.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 2.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 35.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 17.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 75.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 8.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 7.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 7.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 5.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 3.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 5.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 40.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 4.7 | 46.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.8 | 30.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.5 | 30.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.2 | 15.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.7 | 7.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 23.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 10.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 10.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.6 | 7.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.5 | 7.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 19.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 11.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.5 | 9.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 8.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 9.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.4 | 7.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 78.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.4 | 4.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 14.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 4.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 8.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 3.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 9.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 1.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 6.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 8.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 4.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 10.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.2 | 8.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 7.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 10.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 2.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 6.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 8.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 7.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.2 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 2.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |