Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
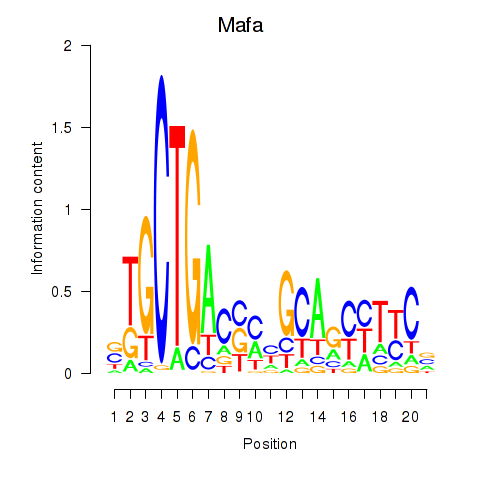
Results for Mafa
Z-value: 0.58
Transcription factors associated with Mafa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mafa
|
ENSRNOG00000007668 | MAF bZIP transcription factor A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafa | rn6_v1_chr7_-_116781766_116781766 | 0.17 | 2.3e-03 | Click! |
Activity profile of Mafa motif
Sorted Z-values of Mafa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_53818818 | 29.24 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr8_+_32018560 | 16.52 |
ENSRNOT00000007358
|
Adamts8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr1_-_174495258 | 16.09 |
ENSRNOT00000052377
ENSRNOT00000086349 |
Scube2
|
signal peptide, CUB domain and EGF like domain containing 2 |
| chr16_-_18766174 | 16.06 |
ENSRNOT00000084813
|
Sftpd
|
surfactant protein D |
| chr10_+_73333119 | 14.90 |
ENSRNOT00000004736
|
Tbx4
|
T-box 4 |
| chr11_+_17538063 | 14.43 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr3_-_117766120 | 13.26 |
ENSRNOT00000056022
|
Fbn1
|
fibrillin 1 |
| chr5_+_173288447 | 11.66 |
ENSRNOT00000091205
ENSRNOT00000067252 |
Mxra8
|
matrix remodeling associated 8 |
| chr10_+_94170766 | 10.79 |
ENSRNOT00000010627
|
Ace
|
angiotensin I converting enzyme |
| chr1_+_272799784 | 10.10 |
ENSRNOT00000016052
|
Ins1
|
insulin 1 |
| chr16_-_69132584 | 10.02 |
ENSRNOT00000017776
|
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr3_+_15560712 | 9.98 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr20_-_21689553 | 9.36 |
ENSRNOT00000038095
|
Tmem26
|
transmembrane protein 26 |
| chr19_-_11341863 | 8.45 |
ENSRNOT00000025694
|
Mt4
|
metallothionein 4 |
| chr3_-_7203420 | 7.60 |
ENSRNOT00000015236
|
Gfi1b
|
growth factor independent 1B transcriptional repressor |
| chr1_-_254735548 | 7.52 |
ENSRNOT00000025258
|
Ankrd1
|
ankyrin repeat domain 1 |
| chr3_+_6430201 | 7.39 |
ENSRNOT00000086352
|
Col5a1
|
collagen type V alpha 1 chain |
| chr1_+_282265370 | 7.30 |
ENSRNOT00000015687
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr10_+_36098051 | 6.89 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr1_-_215858034 | 6.71 |
ENSRNOT00000027656
|
Ins2
|
insulin 2 |
| chr20_-_12938891 | 6.26 |
ENSRNOT00000017141
|
RGD1564149
|
similar to Protein C21orf58 |
| chr11_+_57265732 | 5.82 |
ENSRNOT00000093174
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr20_-_32139789 | 4.94 |
ENSRNOT00000078140
|
Srgn
|
serglycin |
| chr18_+_30172740 | 4.90 |
ENSRNOT00000027340
|
Pcdha4
|
protocadherin alpha 4 |
| chr7_-_12424367 | 4.12 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr20_+_4329811 | 4.09 |
ENSRNOT00000000513
|
Notch4
|
notch 4 |
| chr15_+_105851542 | 4.04 |
ENSRNOT00000086959
|
Rap2a
|
RAS related protein 2a |
| chr1_+_221099998 | 3.71 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr7_-_117267402 | 3.71 |
ENSRNOT00000088945
|
Plec
|
plectin |
| chr12_-_22726982 | 3.68 |
ENSRNOT00000001921
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr7_-_117267803 | 3.49 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr6_+_102392828 | 3.31 |
ENSRNOT00000089162
|
Rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr1_-_145931583 | 3.25 |
ENSRNOT00000016433
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr9_+_16003058 | 2.94 |
ENSRNOT00000081621
ENSRNOT00000021158 |
Ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr13_+_52147555 | 2.72 |
ENSRNOT00000084766
|
Lmod1
|
leiomodin 1 |
| chr20_-_7930929 | 2.71 |
ENSRNOT00000000607
|
Tead3
|
TEA domain transcription factor 3 |
| chr20_+_5509059 | 2.42 |
ENSRNOT00000065349
|
Kifc1
|
kinesin family member C1 |
| chr9_+_16543688 | 2.39 |
ENSRNOT00000021868
|
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr6_-_138909105 | 2.39 |
ENSRNOT00000087855
|
AABR07065656.9
|
|
| chr1_-_224974203 | 2.32 |
ENSRNOT00000065694
|
Tmem179b
|
transmembrane protein 179B |
| chrX_+_22247088 | 2.29 |
ENSRNOT00000079802
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr8_-_116438038 | 2.25 |
ENSRNOT00000023791
|
Gnat1
|
G protein subunit alpha transducin 1 |
| chr18_+_65814026 | 2.24 |
ENSRNOT00000016112
|
Mbd2
|
methyl-CpG binding domain protein 2 |
| chr1_-_225128740 | 2.15 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr1_-_167210919 | 2.11 |
ENSRNOT00000027507
|
Chrna10
|
cholinergic receptor nicotinic alpha 10 subunit |
| chr2_-_27949066 | 2.04 |
ENSRNOT00000081472
ENSRNOT00000022138 |
Nsa2
|
NSA2 ribosome biogenesis homolog |
| chrX_-_10413984 | 2.03 |
ENSRNOT00000039551
ENSRNOT00000091448 |
Ddx3x
|
DEAD-box helicase 3, X-linked |
| chr5_+_26493212 | 2.01 |
ENSRNOT00000061328
|
Triqk
|
triple QxxK/R motif containing |
| chr10_+_83081168 | 1.94 |
ENSRNOT00000035023
|
Tac4
|
tachykinin 4 (hemokinin) |
| chr1_+_245237736 | 1.90 |
ENSRNOT00000035814
|
Vldlr
|
very low density lipoprotein receptor |
| chr1_+_162083030 | 1.84 |
ENSRNOT00000016361
|
Gab2
|
GRB2-associated binding protein 2 |
| chr20_-_5056474 | 1.81 |
ENSRNOT00000076375
ENSRNOT00000037191 ENSRNOT00000076863 |
G6b
|
immunoreceptor tyrosine-based inhibitory motif (ITIM) containing platelet receptor |
| chr1_-_170594168 | 1.80 |
ENSRNOT00000026280
|
Tpp1
|
tripeptidyl peptidase 1 |
| chrX_-_111102464 | 1.80 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr2_+_185524774 | 1.78 |
ENSRNOT00000089338
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr10_+_14828597 | 1.57 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr13_-_74276964 | 1.33 |
ENSRNOT00000005726
|
Tor3a
|
torsin family 3, member A |
| chr7_+_13318533 | 1.17 |
ENSRNOT00000034545
|
LOC691170
|
similar to zinc finger protein 84 (HPF2) |
| chr8_-_107602263 | 1.16 |
ENSRNOT00000017658
|
Esyt3
|
extended synaptotagmin 3 |
| chr6_-_127653124 | 1.05 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr13_-_37287458 | 0.95 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr18_+_44810388 | 0.94 |
ENSRNOT00000021646
ENSRNOT00000089750 |
Hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chrX_+_123913072 | 0.88 |
ENSRNOT00000071376
|
Rhox5
|
reproductive homeobox 5 |
| chr1_-_213987053 | 0.86 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr5_+_3955227 | 0.70 |
ENSRNOT00000009908
|
Msc
|
musculin |
| chr4_-_144318580 | 0.69 |
ENSRNOT00000007591
|
Ssuh2
|
ssu-2 homolog |
| chr1_-_87221826 | 0.66 |
ENSRNOT00000046611
ENSRNOT00000028006 |
Spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr20_+_10438444 | 0.50 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr3_-_60813869 | 0.46 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr8_+_75625174 | 0.42 |
ENSRNOT00000013742
|
Ice2
|
interactor of little elongation complex ELL subunit 2 |
| chr5_-_58113553 | 0.38 |
ENSRNOT00000046589
|
Arid3c
|
AT-rich interaction domain 3C |
| chr10_+_89174684 | 0.31 |
ENSRNOT00000043754
|
Vps25
|
vacuolar protein sorting 25 |
| chr4_+_96252798 | 0.11 |
ENSRNOT00000008947
|
RGD1560028
|
similar to RIKEN cDNA C130060K24 gene |
| chr8_+_42284654 | 0.10 |
ENSRNOT00000043810
|
LOC100911939
|
olfactory receptor 8B3-like |
| chr3_+_176865156 | 0.09 |
ENSRNOT00000019084
|
Zgpat
|
zinc finger CCCH-type and G-patch domain containing |
| chr14_+_114126943 | 0.01 |
ENSRNOT00000041638
ENSRNOT00000006443 ENSRNOT00000006957 |
Rtn4
|
reticulon 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mafa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 3.3 | 10.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 3.2 | 16.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 3.2 | 16.1 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 2.9 | 11.7 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 2.7 | 10.8 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 2.7 | 29.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 2.2 | 6.7 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 2.0 | 10.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.8 | 7.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.6 | 4.9 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.2 | 3.7 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.2 | 3.7 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.2 | 9.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.0 | 4.1 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.0 | 7.6 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.8 | 4.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 5.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.5 | 1.9 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.4 | 7.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.4 | 7.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 2.3 | GO:0051343 | sensory perception of umami taste(GO:0050917) positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.4 | 2.9 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.3 | 2.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.3 | 0.9 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.3 | 2.0 | GO:0042256 | mature ribosome assembly(GO:0042256) protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 4.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 2.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 2.3 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 0.7 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 2.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 0.7 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 6.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 2.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.8 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 14.9 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.1 | 1.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 2.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 3.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.5 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 2.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 2.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 1.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 9.8 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 4.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 29.2 | GO:0032982 | myosin filament(GO:0032982) |
| 1.8 | 7.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.2 | 13.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 12.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 16.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.4 | 5.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 7.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 6.7 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 6.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 8.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 2.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.9 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 1.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 4.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 7.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 7.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 17.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 31.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 10.1 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 2.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 10.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 4.9 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 3.6 | 10.8 | GO:0070573 | bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 1.3 | 29.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 1.2 | 3.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 1.2 | 7.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.6 | 16.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.6 | 1.9 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.6 | 1.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.5 | 7.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 7.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 14.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 2.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 16.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.4 | 1.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 2.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.3 | 0.9 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.3 | 2.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 13.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 10.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 7.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 23.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 4.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 3.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 3.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 2.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 4.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 10.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 4.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 5.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 14.5 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 10.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 16.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 5.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 7.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 7.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 26.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 4.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 8.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 29.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 4.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.3 | 18.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 7.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 10.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 2.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 13.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 7.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 7.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |