Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
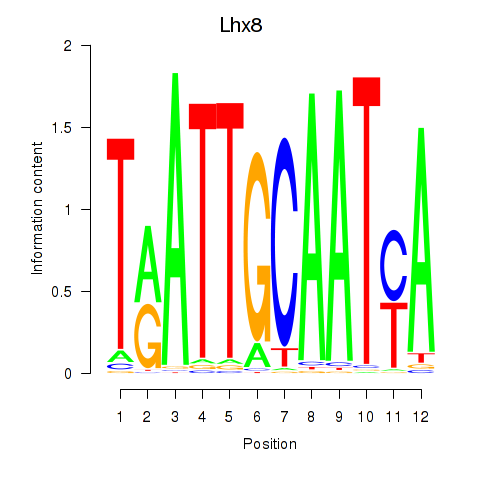
Results for Lhx8
Z-value: 0.73
Transcription factors associated with Lhx8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx8
|
ENSRNOG00000028348 | LIM homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx8 | rn6_v1_chr2_-_260596777_260596777 | 0.35 | 1.8e-10 | Click! |
Activity profile of Lhx8 motif
Sorted Z-values of Lhx8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_48106099 | 38.59 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr14_+_99529284 | 28.72 |
ENSRNOT00000006897
ENSRNOT00000067134 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr3_+_51687809 | 27.52 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chrX_-_56765893 | 27.17 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr3_-_38090526 | 25.65 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr4_-_159192526 | 24.58 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr6_-_14523961 | 24.42 |
ENSRNOT00000071402
|
Nrxn1
|
neurexin 1 |
| chr8_-_59226597 | 21.10 |
ENSRNOT00000088208
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr2_-_57935334 | 20.33 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr2_-_170460754 | 20.32 |
ENSRNOT00000013009
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr1_+_37507276 | 20.14 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chr18_+_30515962 | 19.93 |
ENSRNOT00000027172
|
LOC108348233
|
protocadherin beta-6-like |
| chr18_+_30474947 | 19.18 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr4_+_70252366 | 18.83 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr2_-_181531978 | 18.48 |
ENSRNOT00000072029
|
Npy2r
|
neuropeptide Y receptor Y2 |
| chr5_-_16140896 | 17.75 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr6_-_99870024 | 17.27 |
ENSRNOT00000010043
|
Rab15
|
RAB15, member RAS oncogene family |
| chr11_+_74834050 | 16.88 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chr18_+_27047382 | 16.06 |
ENSRNOT00000027691
ENSRNOT00000090264 |
Apc
|
APC, WNT signaling pathway regulator |
| chr13_+_82072497 | 15.24 |
ENSRNOT00000063810
ENSRNOT00000085135 |
Kifap3
|
kinesin-associated protein 3 |
| chr12_-_10335499 | 13.50 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr6_+_9790422 | 13.27 |
ENSRNOT00000020959
|
Prkce
|
protein kinase C, epsilon |
| chr19_+_320906 | 12.84 |
ENSRNOT00000041664
|
AABR07042611.1
|
|
| chr7_-_82059247 | 12.72 |
ENSRNOT00000087177
|
Rspo2
|
R-spondin 2 |
| chr18_+_63203063 | 12.55 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr10_-_51576376 | 12.24 |
ENSRNOT00000004829
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr2_-_18531210 | 12.19 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr6_+_28515025 | 12.15 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr18_+_29951094 | 11.10 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr8_-_65587658 | 11.07 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr13_+_24823488 | 10.93 |
ENSRNOT00000019907
|
Cdh20
|
cadherin 20 |
| chr18_+_30424814 | 10.63 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr12_-_5773036 | 10.04 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr9_-_63641400 | 9.95 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr2_-_173563273 | 9.94 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr20_+_1749716 | 9.71 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr20_+_1764794 | 9.65 |
ENSRNOT00000075084
|
Olr1736
|
olfactory receptor 1736 |
| chr6_+_95323579 | 9.46 |
ENSRNOT00000007369
|
Pcnx4
|
pecanex homolog 4 (Drosophila) |
| chr3_-_105470475 | 9.41 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr2_-_147819335 | 9.38 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr1_+_42169501 | 8.98 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr9_-_52401660 | 8.83 |
ENSRNOT00000049387
|
Vom1r50
|
vomeronasal 1 receptor 50 |
| chr9_-_65442257 | 8.67 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr18_+_30909490 | 8.11 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr16_+_39909270 | 8.03 |
ENSRNOT00000081994
|
Wdr17
|
WD repeat domain 17 |
| chr8_+_52127632 | 7.75 |
ENSRNOT00000079797
|
Cadm1
|
cell adhesion molecule 1 |
| chr1_-_67094567 | 7.66 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr3_+_80362858 | 7.38 |
ENSRNOT00000021353
ENSRNOT00000086242 |
Lrp4
|
LDL receptor related protein 4 |
| chr6_+_43234526 | 7.17 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr12_+_2140203 | 6.63 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr4_-_150506557 | 6.58 |
ENSRNOT00000076927
|
Zfp248
|
zinc finger protein 248 |
| chr16_-_75637789 | 6.39 |
ENSRNOT00000058029
|
Defb4
|
defensin beta 4 |
| chr2_+_72006099 | 6.29 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr7_-_40316532 | 6.08 |
ENSRNOT00000083347
|
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr16_-_3765917 | 6.02 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr1_+_204959174 | 5.91 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr3_-_77227036 | 5.85 |
ENSRNOT00000072638
|
LOC103691843
|
olfactory receptor 4P4-like |
| chr10_+_90230711 | 5.82 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chrX_-_2657155 | 5.70 |
ENSRNOT00000005630
|
Chst7
|
carbohydrate sulfotransferase 7 |
| chr7_+_2831004 | 5.60 |
ENSRNOT00000029778
|
Rnf41
|
ring finger protein 41 |
| chr6_-_76608864 | 5.56 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr3_-_80000091 | 5.55 |
ENSRNOT00000079394
|
Madd
|
MAP-kinase activating death domain |
| chr1_-_79899648 | 5.42 |
ENSRNOT00000057960
|
AC110846.1
|
|
| chrX_-_138112408 | 5.41 |
ENSRNOT00000077028
|
Frmd7
|
FERM domain containing 7 |
| chr10_-_45297385 | 5.28 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr10_-_52290657 | 5.07 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr7_-_15301382 | 4.99 |
ENSRNOT00000081512
|
Zfp763
|
zinc finger protein 763 |
| chr15_-_11812485 | 4.88 |
ENSRNOT00000030991
ENSRNOT00000007795 |
Nek10
|
NIMA-related kinase 10 |
| chr4_+_160020472 | 4.88 |
ENSRNOT00000078802
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr3_-_101547478 | 4.80 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr15_-_93868292 | 4.67 |
ENSRNOT00000093546
ENSRNOT00000014583 |
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr9_+_16702460 | 4.58 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chr2_+_196145384 | 4.43 |
ENSRNOT00000080966
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr7_-_9465527 | 4.42 |
ENSRNOT00000044882
|
Olr1069
|
olfactory receptor 1069 |
| chr16_-_49820235 | 4.42 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr10_-_13996565 | 4.37 |
ENSRNOT00000016221
ENSRNOT00000052138 |
Tsc2
|
tuberous sclerosis 2 |
| chr1_-_49844547 | 4.24 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr11_-_82810014 | 4.21 |
ENSRNOT00000083539
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chrX_-_77559348 | 4.03 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr4_-_150506406 | 4.02 |
ENSRNOT00000076307
|
Zfp248
|
zinc finger protein 248 |
| chr1_-_103426467 | 3.84 |
ENSRNOT00000045792
|
Mrgprb4
|
MAS-related GPR, member B4 |
| chr1_+_218466289 | 3.75 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr2_-_38110567 | 3.71 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr5_+_144581427 | 3.69 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chr3_-_9936352 | 3.59 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr5_-_137617258 | 3.58 |
ENSRNOT00000071641
|
Olfr1330-ps1
|
olfactory receptor 1330, pseudogene 1 |
| chr6_+_55374984 | 3.32 |
ENSRNOT00000091682
|
Agr3
|
anterior gradient 3, protein disulphide isomerase family member |
| chrX_+_32495809 | 3.31 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr1_-_48891130 | 3.28 |
ENSRNOT00000083884
|
AC135026.1
|
|
| chr13_+_82355886 | 3.18 |
ENSRNOT00000076757
|
Sele
|
selectin E |
| chr1_-_62114672 | 3.14 |
ENSRNOT00000072607
|
AABR07001923.1
|
|
| chr1_+_168489077 | 3.02 |
ENSRNOT00000021191
|
Olr96
|
olfactory receptor 96 |
| chr11_-_71136673 | 3.02 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr7_-_15852930 | 3.01 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chr1_-_242441247 | 2.98 |
ENSRNOT00000068645
|
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chrX_-_119162518 | 2.88 |
ENSRNOT00000073176
|
LOC103694506
|
ubiquitin-conjugating enzyme E2 W |
| chrX_+_71540895 | 2.86 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chrX_-_29825439 | 2.86 |
ENSRNOT00000048155
|
Gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr1_+_219250265 | 2.84 |
ENSRNOT00000024353
|
Doc2g
|
double C2-like domains, gamma |
| chr16_+_32457521 | 2.81 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr20_+_1736377 | 2.77 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr2_-_258997138 | 2.74 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr13_+_82355471 | 2.71 |
ENSRNOT00000030677
|
Sele
|
selectin E |
| chr7_-_140342727 | 2.71 |
ENSRNOT00000089612
|
Ddx23
|
DEAD-box helicase 23 |
| chr8_+_41336340 | 2.64 |
ENSRNOT00000072049
|
Olr1225
|
olfactory receptor 1225 |
| chr1_-_261371508 | 2.62 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr2_+_189857587 | 2.54 |
ENSRNOT00000048214
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr4_+_2593876 | 2.51 |
ENSRNOT00000092992
|
Ube3c
|
ubiquitin protein ligase E3C |
| chr10_-_45812641 | 2.46 |
ENSRNOT00000038951
|
Zfp867
|
zinc finger protein 867 |
| chr9_-_38495126 | 2.44 |
ENSRNOT00000016933
ENSRNOT00000090385 |
Rab23
|
RAB23, member RAS oncogene family |
| chr1_+_70454322 | 2.42 |
ENSRNOT00000072272
|
Olr4
|
olfactory receptor 4 |
| chr11_+_88424414 | 2.40 |
ENSRNOT00000022328
|
Spag6l
|
sperm associated antigen 6-like |
| chr5_-_78183122 | 2.39 |
ENSRNOT00000002083
ENSRNOT00000067076 |
Fkbp15
|
FK506 binding protein 15 |
| chr16_+_6609668 | 2.34 |
ENSRNOT00000021862
|
Tkt
|
transketolase |
| chr1_-_23556241 | 2.34 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr18_+_1866080 | 2.31 |
ENSRNOT00000018646
|
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr10_-_64341066 | 2.30 |
ENSRNOT00000082825
ENSRNOT00000010010 |
Vps53
|
VPS53 GARP complex subunit |
| chr3_+_15038267 | 2.27 |
ENSRNOT00000083134
|
Dab2ip
|
DAB2 interacting protein |
| chr4_-_165828814 | 2.27 |
ENSRNOT00000007481
|
Tas2r105
|
taste receptor, type 2, member 105 |
| chr1_-_168611670 | 2.24 |
ENSRNOT00000021273
|
Olr106
|
olfactory receptor 106 |
| chr5_+_139385429 | 2.23 |
ENSRNOT00000078622
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr13_-_89433815 | 2.20 |
ENSRNOT00000091541
|
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr11_+_38727048 | 2.18 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr7_+_35773928 | 2.17 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr1_+_79899155 | 2.13 |
ENSRNOT00000078503
|
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr3_-_74590459 | 2.08 |
ENSRNOT00000065152
|
Olr535
|
olfactory receptor 535 |
| chr7_+_14643704 | 2.04 |
ENSRNOT00000044642
|
AABR07055875.1
|
|
| chr2_+_62150251 | 2.00 |
ENSRNOT00000016196
|
Zfr
|
zinc finger RNA binding protein |
| chr1_+_221710670 | 1.99 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr2_-_86475096 | 1.99 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr1_+_169616178 | 1.97 |
ENSRNOT00000023170
|
Olr157
|
olfactory receptor 157 |
| chr4_+_123760743 | 1.96 |
ENSRNOT00000013498
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr11_-_70207361 | 1.94 |
ENSRNOT00000002444
|
Muc13
|
mucin 13, cell surface associated |
| chr13_-_31753800 | 1.90 |
ENSRNOT00000050715
|
AC109707.1
|
|
| chr2_+_200076054 | 1.87 |
ENSRNOT00000025327
|
Sec22b
|
SEC22 homolog B, vesicle trafficking protein |
| chr5_-_168123030 | 1.84 |
ENSRNOT00000092747
|
Per3
|
period circadian clock 3 |
| chr2_-_205212681 | 1.83 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chrX_+_31054394 | 1.83 |
ENSRNOT00000037439
|
LOC688526
|
similar to ribonucleic acid binding protein S1 |
| chr17_-_42422053 | 1.81 |
ENSRNOT00000048298
|
Fam65b
|
family with sequence similarity 65, member B |
| chr7_+_121361415 | 1.79 |
ENSRNOT00000067904
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr3_-_10694649 | 1.76 |
ENSRNOT00000037742
|
Gpr107
|
G protein-coupled receptor 107 |
| chrX_-_115426083 | 1.72 |
ENSRNOT00000014756
|
AABR07040947.1
|
|
| chr8_+_13862651 | 1.71 |
ENSRNOT00000014605
|
Taf1d
|
TATA-box binding protein associated factor, RNA polymerase I subunit D |
| chr5_-_137846436 | 1.70 |
ENSRNOT00000046941
|
Olr866
|
olfactory receptor 866 |
| chr4_-_72020642 | 1.70 |
ENSRNOT00000075946
|
Fam115e
|
family with sequence similarity 115, member E |
| chr1_+_107262659 | 1.68 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr9_+_99720856 | 1.66 |
ENSRNOT00000047487
|
Olr1346
|
olfactory receptor 1346 |
| chr17_-_23792353 | 1.55 |
ENSRNOT00000019089
|
Tbc1d7
|
TBC1 domain family, member 7 |
| chr17_+_16333415 | 1.50 |
ENSRNOT00000060550
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr11_+_45751812 | 1.47 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr1_+_1771710 | 1.44 |
ENSRNOT00000080138
ENSRNOT00000073528 |
Nup43
|
nucleoporin 43 |
| chrX_-_915953 | 1.39 |
ENSRNOT00000075264
|
Spaca5
|
sperm acrosome associated 5 |
| chr14_+_22597103 | 1.29 |
ENSRNOT00000048482
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr7_-_20118466 | 1.28 |
ENSRNOT00000080523
|
RGD1565071
|
similar to hypothetical protein 4930509O22 |
| chr10_+_44228800 | 1.24 |
ENSRNOT00000046817
|
Olr1431
|
olfactory receptor 1431 |
| chr7_-_114590119 | 1.23 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr7_-_9630611 | 1.21 |
ENSRNOT00000011176
|
Olr1072
|
olfactory receptor 1072 |
| chr3_-_76696107 | 1.20 |
ENSRNOT00000044692
|
Olr629
|
olfactory receptor 629 |
| chr7_+_23854846 | 1.19 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr7_+_14743008 | 1.17 |
ENSRNOT00000055284
|
AABR07055878.1
|
|
| chr1_+_229889771 | 1.15 |
ENSRNOT00000085941
|
Olr348
|
olfactory receptor 348 |
| chr20_+_4824226 | 1.11 |
ENSRNOT00000001113
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr4_+_1591444 | 1.04 |
ENSRNOT00000078277
|
Olr1245
|
olfactory receptor 1245 |
| chr9_-_23454316 | 1.00 |
ENSRNOT00000072826
|
Cyp2ac1
|
cytochrome P450, family 2, subfamily ac, polypeptide 1 |
| chr12_-_12782139 | 0.99 |
ENSRNOT00000001392
ENSRNOT00000079836 |
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr12_+_16899025 | 0.97 |
ENSRNOT00000001716
|
Psmg3
|
proteasome assembly chaperone 3 |
| chr7_-_143793774 | 0.90 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr1_-_71373605 | 0.90 |
ENSRNOT00000034854
|
Galp
|
galanin-like peptide |
| chr7_+_132378273 | 0.87 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr2_-_198834925 | 0.87 |
ENSRNOT00000089831
|
Nudt17
|
nudix hydrolase 17 |
| chr3_-_74906989 | 0.86 |
ENSRNOT00000071001
|
Olr545
|
olfactory receptor 545 |
| chr17_-_2705123 | 0.84 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr1_-_171045419 | 0.79 |
ENSRNOT00000047239
|
Olr224
|
olfactory receptor 224 |
| chr1_+_86103298 | 0.74 |
ENSRNOT00000014883
|
Vom2r10
|
vomeronasal 2 receptor, 10 |
| chr7_-_130408187 | 0.74 |
ENSRNOT00000015374
|
Chkb
|
choline kinase beta |
| chr5_-_164648328 | 0.73 |
ENSRNOT00000090811
|
Zfp933
|
zinc finger protein 933 |
| chr5_-_157059508 | 0.72 |
ENSRNOT00000022282
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr3_-_103140972 | 0.70 |
ENSRNOT00000052368
|
Olr780
|
olfactory receptor 780 |
| chr10_-_44750464 | 0.66 |
ENSRNOT00000042919
|
Olr1457
|
olfactory receptor 1457 |
| chr10_-_60573129 | 0.65 |
ENSRNOT00000083428
|
Olr1499
|
olfactory receptor 1499 |
| chr10_+_29033989 | 0.65 |
ENSRNOT00000088322
|
Slu7
|
SLU7 homolog, splicing factor |
| chr18_-_71614980 | 0.64 |
ENSRNOT00000032563
|
LOC102548286
|
peroxisomal biogenesis factor 19-like |
| chr17_+_78915604 | 0.63 |
ENSRNOT00000057855
|
Rpp38
|
ribonuclease P/MRP 38 subunit |
| chr9_-_88534710 | 0.60 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr3_-_75733970 | 0.58 |
ENSRNOT00000033519
|
Olr577
|
olfactory receptor 577 |
| chr20_+_4823662 | 0.57 |
ENSRNOT00000090028
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr4_+_88026033 | 0.55 |
ENSRNOT00000074905
|
Vom1r80
|
vomeronasal 1 receptor 80 |
| chr10_+_60974225 | 0.54 |
ENSRNOT00000042349
|
Olr1511
|
olfactory receptor 1511 |
| chr1_-_169892850 | 0.51 |
ENSRNOT00000037868
|
Olr183
|
olfactory receptor 183 |
| chr9_+_79659251 | 0.50 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr2_+_235738416 | 0.50 |
ENSRNOT00000074209
|
Etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr3_-_77365887 | 0.49 |
ENSRNOT00000090742
|
LOC686900
|
similar to olfactory receptor 1198 |
| chrX_+_32232142 | 0.49 |
ENSRNOT00000047354
|
Ca5b
|
carbonic anhydrase 5B |
| chr1_+_65906344 | 0.48 |
ENSRNOT00000068611
|
Vom2r34
|
vomeronasal 2 receptor, 34 |
| chr3_+_78086943 | 0.44 |
ENSRNOT00000047425
|
Olr691
|
olfactory receptor 691 |
| chr1_+_230217215 | 0.44 |
ENSRNOT00000072772
|
LOC687088
|
similar to olfactory receptor 1467 |
| chr8_-_87419564 | 0.43 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr3_+_102859815 | 0.43 |
ENSRNOT00000037906
|
Olr771
|
olfactory receptor 771 |
| chr14_-_87701884 | 0.41 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr1_-_66321642 | 0.40 |
ENSRNOT00000074234
|
AABR07002093.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 59.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 5.7 | 28.7 | GO:0070346 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 5.1 | 20.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 4.9 | 24.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 4.8 | 28.8 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 4.2 | 21.1 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 3.4 | 27.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 3.3 | 13.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 3.1 | 18.5 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 3.1 | 12.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.5 | 10.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 2.0 | 38.6 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 2.0 | 25.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 2.0 | 5.9 | GO:1990168 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 1.8 | 5.5 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 1.5 | 7.7 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 1.4 | 20.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 1.3 | 6.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.3 | 5.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.0 | 15.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.0 | 9.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.9 | 10.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.9 | 4.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) negative regulation of pinocytosis(GO:0048550) |
| 0.8 | 8.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.7 | 2.9 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.7 | 13.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.7 | 2.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.6 | 5.6 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.6 | 81.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.6 | 2.2 | GO:0002884 | immune complex clearance(GO:0002434) negative regulation of hypersensitivity(GO:0002884) |
| 0.5 | 3.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 5.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 2.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.4 | 3.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 4.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.4 | 4.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.3 | 17.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 1.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.3 | 0.9 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.3 | 23.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 2.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.3 | 3.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 12.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.3 | 5.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.0 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.2 | 12.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 2.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 5.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 1.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 4.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 12.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 7.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 2.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 1.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 5.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 2.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 1.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 4.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 2.9 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 4.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 5.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 9.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.3 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 0.2 | GO:0046127 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 0.1 | 4.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 5.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 2.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.3 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.8 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 2.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 16.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 3.0 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 5.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.8 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.9 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 1.8 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 2.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.8 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 38.6 | GO:0097441 | basilar dendrite(GO:0097441) |
| 5.4 | 16.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 3.0 | 15.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.9 | 52.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.5 | 4.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.9 | 6.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.8 | 20.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.8 | 2.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.7 | 7.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.6 | 2.3 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.6 | 1.7 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.6 | 25.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.6 | 24.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.6 | 7.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.5 | 2.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 9.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 2.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 2.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 3.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 2.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 12.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 5.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 2.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 4.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 2.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 2.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 5.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 24.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 9.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.5 | GO:0043564 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 2.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 9.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 24.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 17.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 17.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 21.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 2.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 4.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 43.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 74.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 5.2 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.1 | 5.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 4.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 7.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 10.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 12.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 5.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0000502 | proteasome complex(GO:0000502) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 4.1 | 20.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 3.7 | 18.5 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 3.5 | 24.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 3.3 | 13.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 2.3 | 27.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 2.1 | 21.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.9 | 5.6 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 1.8 | 27.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.4 | 38.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 1.4 | 25.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.3 | 5.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.1 | 4.6 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.1 | 16.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.1 | 24.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.1 | 20.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.0 | 2.9 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.9 | 5.7 | GO:0034481 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) chondroitin sulfotransferase activity(GO:0034481) |
| 0.9 | 2.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.9 | 4.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.8 | 16.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.7 | 3.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.6 | 6.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.6 | 12.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 7.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 5.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 5.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.4 | 2.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.4 | 2.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.4 | 3.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 2.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 15.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 2.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.3 | 1.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 12.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 5.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 4.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 0.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 1.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 4.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 2.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 4.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 3.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 18.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 5.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 4.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 12.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 59.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 7.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.2 | GO:0017113 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 7.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 8.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 5.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 5.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 3.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 6.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 4.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 33.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.8 | 19.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.4 | 10.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 5.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 4.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 9.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 7.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 4.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 5.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 5.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 5.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 10.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 20.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 1.0 | 16.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.9 | 12.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.8 | 27.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 25.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 13.3 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.5 | 9.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.5 | 14.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 24.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 15.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 4.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 5.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 20.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 5.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 6.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.2 | 4.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 9.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 5.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 1.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 17.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 9.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 2.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |