Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Lhx5_Lmx1b_Lhx1
Z-value: 0.85
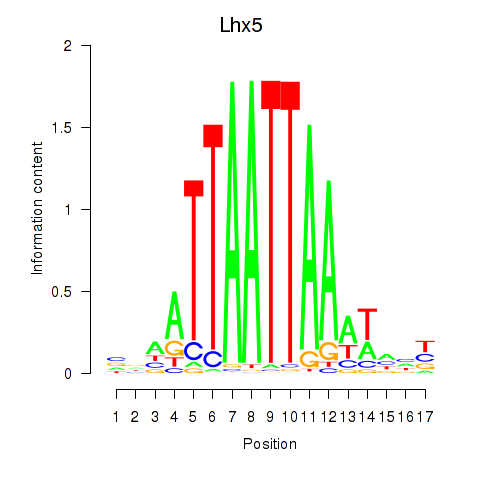
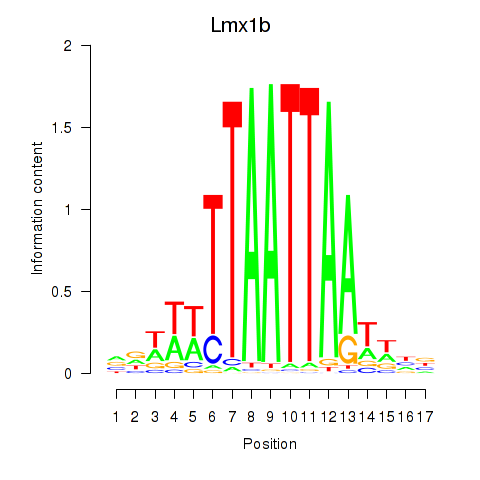
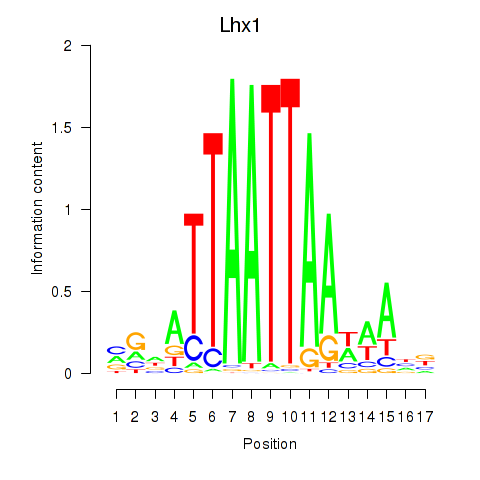
Transcription factors associated with Lhx5_Lmx1b_Lhx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx5
|
ENSRNOG00000001392 | LIM homeobox 5 |
|
Lmx1b
|
ENSRNOG00000017019 | LIM homeobox transcription factor 1 beta |
|
Lhx1
|
ENSRNOG00000002812 | LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx1 | rn6_v1_chr10_-_71849293_71849293 | -0.51 | 6.6e-23 | Click! |
| Lhx5 | rn6_v1_chr12_-_41671437_41671437 | -0.46 | 2.3e-18 | Click! |
| Lmx1b | rn6_v1_chr3_-_12686869_12686869 | -0.20 | 2.3e-04 | Click! |
Activity profile of Lhx5_Lmx1b_Lhx1 motif
Sorted Z-values of Lhx5_Lmx1b_Lhx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_54352270 | 46.38 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr4_+_29535852 | 41.84 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chrX_-_40086870 | 38.01 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr4_-_41212072 | 30.54 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr4_+_180291389 | 29.17 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr3_-_37854561 | 27.77 |
ENSRNOT00000076095
|
Neb
|
nebulin |
| chr17_+_25082056 | 26.52 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr13_-_90074952 | 24.18 |
ENSRNOT00000038006
|
Slamf7
|
SLAM family member 7 |
| chr4_+_163349125 | 23.46 |
ENSRNOT00000084823
|
Klre1
|
killer cell lectin-like receptor, family E, member 1 |
| chr5_+_165724027 | 20.87 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr9_+_53013413 | 17.77 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chrX_+_71342775 | 17.44 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr19_+_27404712 | 17.11 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr20_+_42966140 | 16.70 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr16_+_29674793 | 16.50 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr1_-_101095594 | 15.65 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr6_-_142353308 | 15.31 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chr18_+_32336102 | 14.83 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr9_-_53315915 | 14.65 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr4_+_101882994 | 13.30 |
ENSRNOT00000087773
|
AABR07060963.1
|
|
| chr2_-_54777729 | 13.23 |
ENSRNOT00000082548
|
C7
|
complement C7 |
| chr20_-_5166448 | 12.61 |
ENSRNOT00000076331
|
Aif1
|
allograft inflammatory factor 1 |
| chr4_+_101687327 | 12.22 |
ENSRNOT00000082501
|
AABR07060957.1
|
|
| chr4_+_169147243 | 12.09 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr17_-_18592750 | 11.96 |
ENSRNOT00000065742
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr20_+_3176107 | 11.90 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr1_+_101603222 | 11.63 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr6_+_49825469 | 11.61 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr4_+_169161585 | 11.55 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr8_+_104106740 | 11.48 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr18_+_79773608 | 11.45 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr10_+_53740841 | 11.45 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr4_-_163762434 | 11.25 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr2_-_123972356 | 10.92 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr20_-_4132604 | 10.90 |
ENSRNOT00000077630
ENSRNOT00000048332 |
RT1-Da
|
RT1 class II, locus Da |
| chr11_-_60547201 | 10.32 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr16_-_29936307 | 10.28 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr7_-_69982592 | 10.26 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr19_-_29968424 | 10.05 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr17_-_14679409 | 10.01 |
ENSRNOT00000020704
|
Aspn
|
asporin |
| chr17_+_32082937 | 10.01 |
ENSRNOT00000074775
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr6_-_21112734 | 9.65 |
ENSRNOT00000079819
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr18_+_55666027 | 9.27 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr3_-_118635268 | 9.26 |
ENSRNOT00000086357
|
Atp8b4
|
ATPase phospholipid transporting 8B4 (putative) |
| chr7_-_29233392 | 9.16 |
ENSRNOT00000064241
|
Spic
|
Spi-C transcription factor |
| chr13_+_49005405 | 9.14 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr4_+_93959152 | 9.07 |
ENSRNOT00000058437
|
AABR07060795.1
|
|
| chr2_+_248398917 | 9.05 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr1_+_255040426 | 9.02 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chr8_+_71914867 | 8.89 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr13_-_77821312 | 8.76 |
ENSRNOT00000082110
|
AABR07021544.1
|
|
| chr11_-_60546997 | 8.76 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr19_-_10513349 | 8.69 |
ENSRNOT00000061270
|
Adgrg5
|
adhesion G protein-coupled receptor G5 |
| chr4_+_98481520 | 8.66 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr8_+_5790034 | 8.63 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr6_-_143195445 | 8.62 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr8_-_39551700 | 8.44 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr3_+_20303979 | 8.38 |
ENSRNOT00000058331
|
AABR07051730.1
|
|
| chr2_+_186776644 | 8.34 |
ENSRNOT00000046778
|
Fcrl3
|
Fc receptor-like 3 |
| chr4_+_93791054 | 8.34 |
ENSRNOT00000042300
|
AABR07060788.1
|
|
| chr4_+_155321553 | 8.15 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr17_-_15519282 | 7.97 |
ENSRNOT00000093577
|
Aspnl1
|
asporin-like 1 |
| chr2_-_170301348 | 7.88 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr17_-_15519060 | 7.82 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr20_-_13994794 | 7.63 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr7_+_2459141 | 7.63 |
ENSRNOT00000075681
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr2_+_219598162 | 7.58 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr14_-_77810147 | 7.47 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chrX_+_131381134 | 7.45 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr10_-_896938 | 7.42 |
ENSRNOT00000086392
|
Nde1
|
nudE neurodevelopment protein 1 |
| chr8_+_22559098 | 7.41 |
ENSRNOT00000041091
|
LOC691141
|
hypothetical protein LOC691141 |
| chr13_-_52413681 | 7.36 |
ENSRNOT00000037100
ENSRNOT00000011801 |
Nav1
|
neuron navigator 1 |
| chr6_-_140880070 | 7.20 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr2_+_240642847 | 7.07 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_51498337 | 7.06 |
ENSRNOT00000012487
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr10_+_106812739 | 7.06 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr11_+_15081774 | 7.05 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr3_+_16846412 | 7.02 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr10_+_69423086 | 6.99 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr15_-_95514259 | 6.91 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr9_+_8054466 | 6.81 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr10_-_86930947 | 6.76 |
ENSRNOT00000081440
|
Top2a
|
topoisomerase (DNA) II alpha |
| chrX_+_109996163 | 6.71 |
ENSRNOT00000093349
|
Nrk
|
Nik related kinase |
| chr11_-_28900376 | 6.70 |
ENSRNOT00000061606
|
Krtap16-5
|
keratin associated protein 16-5 |
| chrX_-_77675487 | 6.70 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr3_-_16441030 | 6.63 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chr17_-_43776460 | 6.62 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr7_-_140291620 | 6.61 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr14_+_7949239 | 6.57 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr16_+_2537248 | 6.53 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr12_+_41316764 | 6.45 |
ENSRNOT00000090867
|
Oas3
|
2'-5'-oligoadenylate synthetase 3 |
| chr14_-_34561696 | 6.43 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr3_+_3389612 | 6.39 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr14_+_21177237 | 6.18 |
ENSRNOT00000004866
|
Jchain
|
immunoglobulin joining chain |
| chr1_+_185863043 | 6.04 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr13_-_83457888 | 5.94 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr16_+_72010106 | 5.92 |
ENSRNOT00000058330
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr13_-_50916982 | 5.84 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chrX_+_110014252 | 5.82 |
ENSRNOT00000093611
|
Nrk
|
Nik related kinase |
| chrX_+_96863891 | 5.76 |
ENSRNOT00000085665
|
AABR07040284.1
|
|
| chr10_-_51778939 | 5.74 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr10_-_85725429 | 5.73 |
ENSRNOT00000005471
|
Rpl23
|
ribosomal protein L23 |
| chr4_+_103495993 | 5.73 |
ENSRNOT00000072325
|
AABR07061068.1
|
|
| chr1_+_260093641 | 5.70 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr10_-_29026002 | 5.58 |
ENSRNOT00000005070
|
Pttg1
|
pituitary tumor-transforming 1 |
| chr1_+_22332090 | 5.55 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr2_+_187447501 | 5.47 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr6_-_140485913 | 5.46 |
ENSRNOT00000048463
|
AABR07065768.1
|
|
| chr1_+_253221812 | 5.32 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr4_+_101531378 | 5.30 |
ENSRNOT00000087546
|
AABR07060944.1
|
|
| chr7_-_123088279 | 5.28 |
ENSRNOT00000071998
|
Tob2
|
transducer of ERBB2, 2 |
| chr3_-_165537940 | 5.23 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr15_+_32828165 | 5.21 |
ENSRNOT00000060253
|
AABR07017902.1
|
|
| chr8_-_43304560 | 5.21 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr1_-_78180216 | 5.19 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr6_+_138432550 | 5.18 |
ENSRNOT00000056829
|
Adam6
|
a disintegrin and metallopeptidase domain 6 |
| chr5_-_162751128 | 5.10 |
ENSRNOT00000068281
|
RGD1559644
|
similar to novel protein similar to esterases |
| chr13_-_30800451 | 5.08 |
ENSRNOT00000046791
|
Rpl21
|
ribosomal protein L21 |
| chr4_+_102262007 | 4.99 |
ENSRNOT00000079328
|
AABR07060992.2
|
|
| chr13_-_61306939 | 4.92 |
ENSRNOT00000086610
|
Rgs21
|
regulator of G-protein signaling 21 |
| chr11_+_54619129 | 4.83 |
ENSRNOT00000059924
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr4_+_156253079 | 4.81 |
ENSRNOT00000013536
|
Clec4d
|
C-type lectin domain family 4, member D |
| chr7_+_16404755 | 4.79 |
ENSRNOT00000044977
|
LOC108349290
|
olfactory receptor 6C70-like |
| chr4_+_101949285 | 4.79 |
ENSRNOT00000058446
|
AABR07060963.3
|
|
| chr11_-_62451149 | 4.76 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_+_126766967 | 4.75 |
ENSRNOT00000052296
|
Cox8c
|
cytochrome c oxidase subunit 8C |
| chr1_-_3849080 | 4.72 |
ENSRNOT00000045301
|
RGD1560633
|
similar to ribosomal protein L27a |
| chr3_+_8430829 | 4.71 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr3_+_148327965 | 4.71 |
ENSRNOT00000010851
ENSRNOT00000088481 |
Tpx2
|
TPX2, microtubule nucleation factor |
| chr1_-_166919302 | 4.70 |
ENSRNOT00000026925
|
Folr2
|
folate receptor beta |
| chrX_+_1543244 | 4.66 |
ENSRNOT00000071634
|
AABR07036746.1
|
|
| chr11_-_82366505 | 4.66 |
ENSRNOT00000041326
|
RGD1559972
|
similar to ribosomal protein L27a |
| chr14_-_24123253 | 4.62 |
ENSRNOT00000002743
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chrX_+_1600790 | 4.62 |
ENSRNOT00000072444
|
AC120727.1
|
|
| chr8_+_107859366 | 4.51 |
ENSRNOT00000039690
|
A4gnt
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr20_-_5020150 | 4.30 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr4_-_66955732 | 4.28 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr1_+_101397828 | 4.28 |
ENSRNOT00000028189
|
Kcna7
|
potassium voltage-gated channel subfamily A member 7 |
| chr17_+_52755621 | 4.22 |
ENSRNOT00000089943
|
Psme1-ps1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha), pseudogene 1 |
| chr7_-_139649286 | 4.21 |
ENSRNOT00000080056
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr14_-_46524476 | 4.18 |
ENSRNOT00000079005
|
LOC257642
|
rRNA promoter binding protein |
| chr10_+_61685645 | 4.18 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chrX_-_106607352 | 4.11 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr2_-_147392062 | 4.11 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr4_+_22898527 | 4.08 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr12_-_5685448 | 4.07 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr3_-_94808861 | 4.05 |
ENSRNOT00000038464
|
Prrg4
|
proline rich and Gla domain 4 |
| chr1_-_63684189 | 4.03 |
ENSRNOT00000085651
|
Lilrc2
|
leukocyte immunoglobulin-like receptor, subfamily C, member 2 |
| chr17_-_46115004 | 4.02 |
ENSRNOT00000087838
|
Aoah
|
acyloxyacyl hydrolase |
| chr5_-_154438361 | 4.00 |
ENSRNOT00000085003
|
AC141344.2
|
|
| chr1_+_128637049 | 3.92 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr9_-_80295446 | 3.92 |
ENSRNOT00000023769
|
Tnp1
|
transition protein 1 |
| chr20_+_3155652 | 3.84 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr11_-_62067655 | 3.79 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_119327822 | 3.78 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr14_+_82603671 | 3.78 |
ENSRNOT00000073177
|
Nkx1-1
|
NK1 homeobox 1 |
| chr6_+_139428999 | 3.78 |
ENSRNOT00000084482
|
AABR07065693.2
|
|
| chr14_-_44375804 | 3.75 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr3_+_112346627 | 3.73 |
ENSRNOT00000074392
|
Snap23
|
synaptosomal-associated protein 23 |
| chr10_+_107502695 | 3.65 |
ENSRNOT00000038088
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr15_+_108318664 | 3.64 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr4_-_167202106 | 3.61 |
ENSRNOT00000038581
|
Tas2r140
|
taste receptor, type 2, member 140 |
| chr4_+_162292305 | 3.57 |
ENSRNOT00000010098
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr7_-_139542461 | 3.56 |
ENSRNOT00000089425
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr8_+_117117430 | 3.56 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr20_+_4357733 | 3.49 |
ENSRNOT00000000509
|
Pbx2
|
PBX homeobox 2 |
| chr10_-_74679858 | 3.45 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr5_+_36566783 | 3.41 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr7_+_42304534 | 3.41 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr16_+_2634603 | 3.33 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr1_-_278042312 | 3.32 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr12_-_40590361 | 3.30 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr4_-_103761881 | 3.30 |
ENSRNOT00000084103
|
AABR07061087.1
|
|
| chr1_-_80835701 | 3.29 |
ENSRNOT00000064305
|
PVR
|
poliovirus receptor |
| chr13_-_91735361 | 3.25 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr8_-_39460844 | 3.16 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr11_-_43022565 | 3.15 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr1_-_88162583 | 3.12 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chr14_+_23405717 | 3.10 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr18_-_55891710 | 3.09 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr11_+_34865532 | 2.99 |
ENSRNOT00000050342
|
Dyrk1a
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr10_-_87564327 | 2.96 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr8_+_100260049 | 2.95 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr4_+_31229913 | 2.95 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr1_+_279633671 | 2.93 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr7_+_40318490 | 2.92 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr2_-_60657712 | 2.91 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr7_-_134482607 | 2.90 |
ENSRNOT00000081104
|
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr3_-_4341771 | 2.85 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr4_+_101909389 | 2.82 |
ENSRNOT00000086458
|
AABR07060963.2
|
|
| chrX_-_124252447 | 2.78 |
ENSRNOT00000061546
|
Rhox12
|
reproductive homeobox 12 |
| chr4_-_169036950 | 2.78 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr1_+_21613148 | 2.76 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr15_-_57651041 | 2.76 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr5_+_33097654 | 2.69 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr16_+_71629525 | 2.67 |
ENSRNOT00000035347
ENSRNOT00000088462 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr18_+_30869628 | 2.67 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr1_+_93242050 | 2.67 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr16_-_75518336 | 2.63 |
ENSRNOT00000058037
|
Defa11
|
defensin alpha 11 |
| chr3_+_38367556 | 2.60 |
ENSRNOT00000049144
|
RGD1559995
|
similar to developmental pluripotency associated 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx5_Lmx1b_Lhx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.5 | GO:0002838 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 7.0 | 20.9 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 4.2 | 12.6 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 4.0 | 11.9 | GO:0032639 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 3.4 | 17.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 3.3 | 16.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 3.0 | 9.1 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 3.0 | 8.9 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 2.9 | 20.4 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 2.4 | 16.7 | GO:1900020 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 2.1 | 12.5 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 1.8 | 10.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 1.8 | 10.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.8 | 5.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.8 | 7.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 1.7 | 7.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.7 | 19.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 1.7 | 10.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 1.7 | 46.4 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 1.6 | 3.3 | GO:0045425 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.5 | 9.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.5 | 7.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 1.4 | 25.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.4 | 6.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 1.4 | 20.5 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 1.3 | 6.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 1.2 | 3.6 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 1.1 | 6.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.1 | 6.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.1 | 7.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.1 | 6.6 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 1.1 | 3.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 1.1 | 13.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 1.0 | 8.3 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 1.0 | 6.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.0 | 4.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.0 | 6.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.0 | 7.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.9 | 2.8 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.9 | 11.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.9 | 4.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.9 | 3.4 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.8 | 5.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.8 | 13.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.8 | 3.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.6 | 5.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.6 | 6.4 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.6 | 1.9 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.6 | 23.5 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.6 | 1.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.6 | 10.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.6 | 4.7 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.6 | 2.3 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.6 | 7.9 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.6 | 3.9 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.6 | 3.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 3.7 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 1.5 | GO:0034034 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.5 | 2.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.5 | 5.8 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.5 | 1.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.5 | 1.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.5 | 1.8 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.5 | 2.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.5 | 5.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.4 | 4.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.4 | 4.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 3.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.4 | 1.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.4 | 2.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.4 | 1.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 7.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.4 | 1.4 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.4 | 8.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.4 | 3.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.3 | 1.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 1.7 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.3 | 15.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 1.6 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.3 | 2.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.3 | 0.9 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.3 | 2.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.3 | 7.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 5.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 1.4 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 1.3 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.3 | 0.8 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.3 | 7.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.3 | 7.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 2.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 3.7 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 5.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 1.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 11.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 0.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 3.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 3.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.9 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 1.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 5.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 21.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.2 | 12.0 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.2 | 1.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 34.5 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.2 | 2.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 2.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 10.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 0.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 3.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 2.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.2 | 0.8 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 4.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 9.2 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.1 | 0.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 1.4 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 2.0 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.1 | 0.3 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 1.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 4.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 2.9 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 1.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 3.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.4 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 1.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 1.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 3.8 | GO:0043467 | regulation of generation of precursor metabolites and energy(GO:0043467) |
| 0.1 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.8 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 4.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.9 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.8 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 2.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 4.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.9 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 4.4 | GO:0044264 | cellular polysaccharide metabolic process(GO:0044264) |
| 0.1 | 4.2 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 2.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 1.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 2.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.5 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 2.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 1.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 23.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.7 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 0.1 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 46.4 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 7.6 | 38.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 4.2 | 16.7 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 4.0 | 11.9 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 2.4 | 7.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 2.3 | 9.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 1.7 | 6.8 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.5 | 6.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.5 | 29.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.3 | 11.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.2 | 8.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.1 | 5.3 | GO:0070938 | contractile ring(GO:0070938) |
| 1.0 | 3.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.9 | 6.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.9 | 25.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.7 | 8.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.7 | 3.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.7 | 10.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 10.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.7 | 4.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.6 | 7.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.5 | 2.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.5 | 12.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 3.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 11.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 9.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 2.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 29.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 4.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 4.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 43.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 4.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 4.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 11.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 6.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 1.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 2.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 12.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 1.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 2.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 1.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 2.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 49.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 5.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 7.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 7.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 5.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 31.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 4.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 5.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 7.9 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 2.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 3.4 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 20.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 10.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 9.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 4.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.5 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 3.9 | 27.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 3.3 | 10.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 2.6 | 7.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.3 | 9.0 | GO:0019002 | GMP binding(GO:0019002) |
| 2.2 | 10.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.7 | 5.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.7 | 6.8 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.6 | 6.4 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.6 | 7.8 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 1.5 | 7.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.3 | 6.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.3 | 6.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.3 | 7.6 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 1.2 | 6.2 | GO:0019862 | IgA binding(GO:0019862) |
| 1.2 | 26.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.2 | 4.7 | GO:0051870 | methotrexate binding(GO:0051870) |
| 1.2 | 7.0 | GO:0031727 | CCR1 chemokine receptor binding(GO:0031726) CCR2 chemokine receptor binding(GO:0031727) |
| 1.1 | 5.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.1 | 25.8 | GO:0015288 | porin activity(GO:0015288) |
| 1.1 | 6.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.1 | 5.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.9 | 4.7 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.9 | 2.8 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.9 | 2.6 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.8 | 14.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.8 | 2.4 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.8 | 2.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.7 | 6.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.7 | 12.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.6 | 41.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.6 | 10.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.5 | 3.3 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.5 | 3.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.5 | 3.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 7.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.5 | 3.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.5 | 1.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 5.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.4 | 4.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 2.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.4 | 2.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.4 | 11.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 1.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.4 | 1.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 5.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 3.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.3 | 0.9 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.3 | 9.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 17.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 1.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.3 | 0.8 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.3 | 9.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 0.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 2.7 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 2.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 4.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 2.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 5.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 11.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 24.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 3.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 1.7 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 16.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 3.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 16.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 4.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 4.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 20.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 5.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 23.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 9.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 10.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 4.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 8.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 9.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 6.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 16.9 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 4.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 5.5 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 23.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.0 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 3.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 3.7 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 30.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 6.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 10.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 12.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.3 | 7.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 7.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 8.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 12.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 14.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 11.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 11.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 6.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 7.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 5.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 6.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 6.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 4.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 16.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.3 | 7.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.1 | 14.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.8 | 11.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.7 | 27.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 6.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 6.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.4 | 9.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.4 | 6.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 3.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.4 | 3.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 4.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.3 | 3.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 5.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.3 | 15.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.3 | 19.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.3 | 7.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.3 | 7.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 9.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 18.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 2.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 9.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 12.7 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 5.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 3.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.2 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 4.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.7 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |