Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
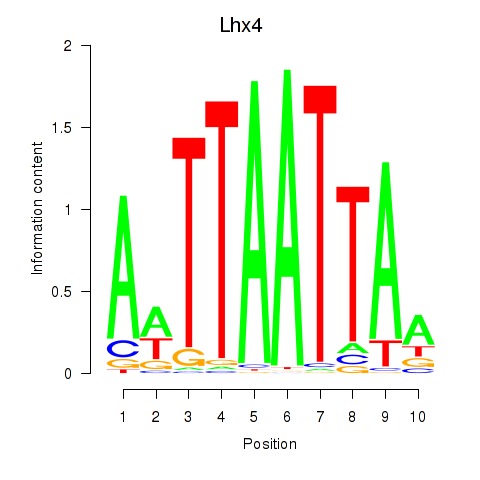
Results for Lhx4
Z-value: 0.54
Transcription factors associated with Lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx4
|
ENSRNOG00000003595 | LIM homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx4 | rn6_v1_chr13_-_73390393_73390543 | -0.34 | 2.4e-10 | Click! |
Activity profile of Lhx4 motif
Sorted Z-values of Lhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_29535852 | 21.07 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr10_+_54352270 | 18.08 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr16_+_29674793 | 16.95 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr17_+_25082056 | 16.51 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr6_-_95934296 | 14.93 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr3_-_37803112 | 13.09 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chrX_-_40086870 | 12.61 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr6_-_60124274 | 11.03 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr4_-_120559078 | 10.40 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr2_-_227411964 | 9.77 |
ENSRNOT00000019931
|
Synpo2
|
synaptopodin 2 |
| chr6_-_23291568 | 9.51 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr20_-_4542073 | 9.27 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chrX_+_156463953 | 8.85 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr14_-_77810147 | 8.68 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr1_+_101603222 | 8.49 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr10_-_51778939 | 8.43 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr16_-_29936307 | 8.35 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr5_-_115387377 | 8.33 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chrX_+_109996163 | 7.91 |
ENSRNOT00000093349
|
Nrk
|
Nik related kinase |
| chr11_+_15081774 | 7.78 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr3_-_7498555 | 7.30 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr17_-_15467320 | 7.12 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr6_-_128149220 | 6.82 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr16_+_2634603 | 6.63 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr2_+_54466280 | 6.35 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr14_+_37116492 | 6.20 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr4_+_57925323 | 6.09 |
ENSRNOT00000085798
|
Cpa5
|
carboxypeptidase A5 |
| chr7_-_69982592 | 5.97 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr15_-_29548400 | 5.93 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr7_-_145062956 | 5.82 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr7_-_73130740 | 5.73 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr18_+_55666027 | 5.54 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr5_+_165724027 | 5.33 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr17_-_69862110 | 5.31 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr3_-_127500709 | 5.30 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr14_+_76732650 | 5.29 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr1_+_279633671 | 5.25 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr10_+_64930023 | 5.20 |
ENSRNOT00000071102
|
AABR07030053.1
|
|
| chr14_-_24123253 | 5.11 |
ENSRNOT00000002743
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chr2_+_193627243 | 4.85 |
ENSRNOT00000082934
|
AABR07012331.1
|
|
| chr10_+_56524468 | 4.81 |
ENSRNOT00000022041
|
Gps2
|
G protein pathway suppressor 2 |
| chr4_+_169147243 | 4.77 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr1_-_104973648 | 4.75 |
ENSRNOT00000019739
|
Dbx1
|
developing brain homeobox 1 |
| chr10_+_45659143 | 4.67 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr16_-_24951612 | 4.64 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr4_+_169161585 | 4.62 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr1_+_255040426 | 4.60 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chr11_-_60547201 | 4.52 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr15_+_110114148 | 4.43 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr8_-_39551700 | 4.38 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr7_-_96464049 | 4.30 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr10_+_94471476 | 4.19 |
ENSRNOT00000088697
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr3_-_166993940 | 4.18 |
ENSRNOT00000034669
|
Zfp217
|
zinc finger protein 217 |
| chr3_+_70327193 | 4.15 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr2_+_127549331 | 4.11 |
ENSRNOT00000093416
|
Slc25a31
|
solute carrier family 25 member 31 |
| chr16_+_71889235 | 4.08 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr13_-_90074952 | 3.94 |
ENSRNOT00000038006
|
Slamf7
|
SLAM family member 7 |
| chr1_-_88162583 | 3.94 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chr4_-_148437961 | 3.93 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr4_-_58250798 | 3.92 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr13_+_26903052 | 3.91 |
ENSRNOT00000003625
|
Serpinb5
|
serpin family B member 5 |
| chr8_-_113689681 | 3.87 |
ENSRNOT00000056435
|
LOC688828
|
similar to Nucleoside diphosphate-linked moiety X motif 16 (Nudix motif 16) |
| chr1_+_128637049 | 3.78 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr3_-_166994286 | 3.73 |
ENSRNOT00000081593
|
Zfp217
|
zinc finger protein 217 |
| chr10_-_88000423 | 3.73 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr19_-_29968424 | 3.71 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_+_98414226 | 3.68 |
ENSRNOT00000090785
|
Siglecl1
|
SIGLEC family like 1 |
| chr1_-_276228574 | 3.65 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr13_+_49005405 | 3.63 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr3_+_8430829 | 3.62 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chrX_-_111191932 | 3.45 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr4_-_10995792 | 3.45 |
ENSRNOT00000078733
|
AABR07059243.1
|
|
| chr11_-_60546997 | 3.38 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr3_+_38367556 | 3.35 |
ENSRNOT00000049144
|
RGD1559995
|
similar to developmental pluripotency associated 5 |
| chr13_+_27449934 | 3.34 |
ENSRNOT00000003409
|
Serpinb2
|
serpin family B member 2 |
| chr14_-_81053905 | 3.20 |
ENSRNOT00000045068
ENSRNOT00000040215 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_+_128614138 | 3.19 |
ENSRNOT00000076227
ENSRNOT00000078707 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr14_-_21758788 | 3.16 |
ENSRNOT00000038520
|
2310003L06Rik
|
RIKEN cDNA 2310003L06 gene |
| chr13_-_91735361 | 3.15 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr2_-_61692487 | 3.14 |
ENSRNOT00000078544
|
LOC499541
|
LRRGT00045 |
| chr1_+_101599018 | 3.13 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chr3_+_76890792 | 3.12 |
ENSRNOT00000087552
|
Olr639
|
olfactory receptor 639 |
| chr1_-_78180216 | 3.09 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr8_-_104155775 | 3.09 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr1_-_101095594 | 2.94 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr18_-_43945273 | 2.92 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr18_+_45023932 | 2.91 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr14_-_44078897 | 2.89 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr5_+_16845631 | 2.80 |
ENSRNOT00000047889
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_+_79021872 | 2.78 |
ENSRNOT00000012677
ENSRNOT00000067125 |
Fam221a
|
family with sequence similarity 221, member A |
| chr1_-_94404211 | 2.77 |
ENSRNOT00000019463
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr6_-_86223052 | 2.73 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr11_+_61605937 | 2.67 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
| chrX_+_23414354 | 2.66 |
ENSRNOT00000031235
|
Cldn34a
|
claudin 34A |
| chr4_-_85314672 | 2.64 |
ENSRNOT00000033672
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr1_-_14117021 | 2.62 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr1_+_166893734 | 2.60 |
ENSRNOT00000026702
|
Phox2a
|
paired-like homeobox 2a |
| chr1_-_173764246 | 2.54 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chrX_-_106607352 | 2.54 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr5_+_119097715 | 2.53 |
ENSRNOT00000045987
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr1_-_128287151 | 2.52 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr6_-_140880070 | 2.51 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr8_-_43304560 | 2.49 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr3_+_3389612 | 2.48 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr1_+_93242050 | 2.46 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr4_+_9882904 | 2.46 |
ENSRNOT00000016909
|
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr20_+_3176107 | 2.44 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr13_-_84795070 | 2.44 |
ENSRNOT00000076083
ENSRNOT00000029706 |
Fmo9
|
flavin containing monooxygenase 9 |
| chr7_-_140291620 | 2.40 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr17_+_63635086 | 2.39 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr10_+_75365822 | 2.38 |
ENSRNOT00000055705
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr20_-_13994794 | 2.38 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr13_-_83425641 | 2.33 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr8_+_5790034 | 2.32 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr2_-_185005572 | 2.29 |
ENSRNOT00000093291
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr5_+_48313599 | 2.28 |
ENSRNOT00000081825
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr3_-_16441030 | 2.27 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chr20_-_7943575 | 2.27 |
ENSRNOT00000066897
|
Tulp1
|
tubby like protein 1 |
| chr4_-_180234804 | 2.26 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr6_-_143195445 | 2.26 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr8_-_39460844 | 2.21 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr11_-_82366505 | 2.19 |
ENSRNOT00000041326
|
RGD1559972
|
similar to ribosomal protein L27a |
| chr8_+_104106740 | 2.16 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr3_-_154627257 | 2.14 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr1_-_56683731 | 2.12 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr10_+_61685645 | 2.11 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr9_+_8054466 | 2.11 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr3_-_90751055 | 2.11 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr20_+_4357733 | 2.11 |
ENSRNOT00000000509
|
Pbx2
|
PBX homeobox 2 |
| chr20_-_5166448 | 2.08 |
ENSRNOT00000076331
|
Aif1
|
allograft inflammatory factor 1 |
| chr5_+_29622281 | 2.08 |
ENSRNOT00000012377
|
Nbn
|
nibrin |
| chr9_+_94324793 | 2.08 |
ENSRNOT00000092493
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr4_-_172063391 | 2.07 |
ENSRNOT00000010158
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr18_+_35384743 | 2.07 |
ENSRNOT00000076143
ENSRNOT00000074593 |
LOC100911797
|
serine protease inhibitor Kazal-type 5-like |
| chr8_-_78397123 | 2.06 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr2_+_127525285 | 2.06 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr18_-_36322320 | 2.06 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
| chr2_-_248789508 | 2.05 |
ENSRNOT00000090705
|
Pkn2
|
protein kinase N2 |
| chr10_-_13446135 | 2.03 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr1_-_3849080 | 2.00 |
ENSRNOT00000045301
|
RGD1560633
|
similar to ribosomal protein L27a |
| chr5_-_162751128 | 1.96 |
ENSRNOT00000068281
|
RGD1559644
|
similar to novel protein similar to esterases |
| chrX_-_124252447 | 1.96 |
ENSRNOT00000061546
|
Rhox12
|
reproductive homeobox 12 |
| chr15_+_27438853 | 1.96 |
ENSRNOT00000011636
|
Olr1622
|
olfactory receptor 1622 |
| chr1_+_212281237 | 1.95 |
ENSRNOT00000075049
|
Utf1
|
undifferentiated embryonic cell transcription factor 1 |
| chr1_+_127802978 | 1.94 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr1_+_79631668 | 1.92 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr11_+_88095170 | 1.88 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr3_-_79090636 | 1.85 |
ENSRNOT00000084116
|
AABR07052846.1
|
|
| chr12_-_23727535 | 1.85 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr7_+_120923274 | 1.84 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr2_+_80269661 | 1.82 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chrX_+_144994139 | 1.82 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr5_-_162442968 | 1.81 |
ENSRNOT00000048366
|
Pramef17
|
PRAME family member 17 |
| chr16_-_49453394 | 1.81 |
ENSRNOT00000041617
|
Lrp2bp
|
Lrp2 binding protein |
| chr9_-_24467892 | 1.80 |
ENSRNOT00000060803
|
Defb18
|
defensin beta 18 |
| chr6_+_49825469 | 1.78 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr10_-_87564327 | 1.77 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr2_+_205160405 | 1.77 |
ENSRNOT00000035605
|
Tspan2
|
tetraspanin 2 |
| chr18_+_65285318 | 1.75 |
ENSRNOT00000020431
|
Tcf4
|
transcription factor 4 |
| chr10_-_87503591 | 1.75 |
ENSRNOT00000037980
|
Krtap1-1
|
keratin associated protein 1-1 |
| chr12_+_48677905 | 1.74 |
ENSRNOT00000083196
|
1700069L16Rik
|
RIKEN cDNA 1700069L16 gene |
| chr3_+_77337698 | 1.72 |
ENSRNOT00000041690
|
LOC103690075
|
olfactory receptor 4P4-like |
| chr9_-_42839837 | 1.69 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr11_-_62396909 | 1.68 |
ENSRNOT00000093596
ENSRNOT00000093179 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_101819478 | 1.66 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr11_-_32550539 | 1.64 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr4_-_56786754 | 1.59 |
ENSRNOT00000050795
|
Kcp
|
kielin/chordin-like protein |
| chr11_+_47061354 | 1.59 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr1_+_185863043 | 1.56 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr1_-_90520344 | 1.56 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr3_+_100769839 | 1.56 |
ENSRNOT00000077703
|
Bdnf
|
brain-derived neurotrophic factor |
| chr9_-_121972055 | 1.56 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr2_-_157759819 | 1.53 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chrX_+_131381134 | 1.51 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr7_-_107203897 | 1.50 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr3_+_31802999 | 1.49 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
| chr5_+_50381244 | 1.47 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr2_-_181900856 | 1.46 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr1_+_74251167 | 1.44 |
ENSRNOT00000040441
|
Vom2r29
|
vomeronasal 2 receptor, 29 |
| chr16_+_84465656 | 1.44 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr2_-_198706428 | 1.43 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr4_+_103495993 | 1.42 |
ENSRNOT00000072325
|
AABR07061068.1
|
|
| chr20_+_42966140 | 1.41 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr4_+_28972434 | 1.39 |
ENSRNOT00000014219
|
Gngt1
|
G protein subunit gamma transducin 1 |
| chr3_+_77289805 | 1.38 |
ENSRNOT00000040581
|
LOC100910858
|
olfactory receptor 4P4-like |
| chr1_+_72380711 | 1.37 |
ENSRNOT00000022236
|
Fiz1
|
FLT3-interacting zinc finger 1 |
| chr3_+_17889972 | 1.36 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr11_+_18454144 | 1.32 |
ENSRNOT00000072550
|
AABR07033357.1
|
|
| chr1_-_37957400 | 1.30 |
ENSRNOT00000085560
ENSRNOT00000079626 ENSRNOT00000071889 |
LOC102549842
|
zinc finger protein 91-like |
| chr1_-_169973241 | 1.29 |
ENSRNOT00000023315
|
Olr191-ps
|
olfactory receptor pseudogene 191 |
| chr1_+_198214797 | 1.28 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr10_+_67810810 | 1.27 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr10_+_112056994 | 1.27 |
ENSRNOT00000073032
|
Vom2r65
|
vomeronasal 2 receptor, 65 |
| chr7_+_122160171 | 1.27 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr16_-_49522341 | 1.25 |
ENSRNOT00000081642
|
LOC100911140
|
coiled-coil domain-containing protein 110-like |
| chr16_+_48513432 | 1.25 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr1_-_813517 | 1.23 |
ENSRNOT00000041332
|
Vom2r5
|
vomeronasal 2 receptor, 5 |
| chr15_-_82581916 | 1.22 |
ENSRNOT00000057891
|
RGD1560069
|
similar to ribosomal protein L27 |
| chr7_-_16010574 | 1.22 |
ENSRNOT00000051296
|
Olr996
|
olfactory receptor 996 |
| chr1_+_230604386 | 1.21 |
ENSRNOT00000040608
|
Olr375
|
olfactory receptor 375 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 14.9 | GO:2000729 | response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 2.8 | 8.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 2.1 | 6.4 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 1.8 | 5.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.8 | 8.9 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 1.3 | 5.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 1.3 | 7.9 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 1.1 | 4.3 | GO:1900623 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) positive regulation of hyaluronan biosynthetic process(GO:1900127) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.1 | 3.2 | GO:0032752 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.0 | 3.1 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.0 | 3.9 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.9 | 6.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.9 | 9.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.9 | 7.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.9 | 2.6 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.8 | 2.4 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.7 | 6.6 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.7 | 2.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.7 | 2.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.7 | 2.6 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.7 | 8.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.6 | 18.1 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.6 | 2.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.6 | 2.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.6 | 6.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.5 | 3.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.5 | 9.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.5 | 1.9 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.4 | 8.7 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.4 | 2.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.4 | 2.4 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 0.4 | 2.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.4 | 4.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.3 | 1.0 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 1.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.3 | 1.6 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 2.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 1.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.2 | 0.7 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.2 | 3.9 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 2.9 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 3.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 4.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 1.1 | GO:0070666 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.2 | 1.1 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 2.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.2 | 1.0 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 1.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 1.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.2 | 3.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 1.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 1.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 3.9 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.2 | 3.7 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 4.3 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 1.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 4.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.4 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 9.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.8 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 2.6 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 7.1 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.7 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.7 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 1.3 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 3.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 1.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 2.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 2.5 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 2.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 12.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 1.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 2.3 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 1.0 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.1 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 2.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 4.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 1.8 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 2.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 2.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 2.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 2.1 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.2 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 27.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.8 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 2.5 | 12.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.0 | 6.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.9 | 6.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 2.4 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.6 | 3.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 3.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 12.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.4 | 3.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 1.4 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.3 | 2.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 8.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 2.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 2.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 2.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 4.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 22.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 2.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 1.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 3.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 37.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.8 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 9.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 8.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 5.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 22.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 3.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 7.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 6.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 5.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 8.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.3 | 5.3 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 1.3 | 3.9 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 1.2 | 3.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 1.0 | 3.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 1.0 | 3.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.0 | 2.9 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.9 | 4.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 18.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.8 | 3.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.8 | 5.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.7 | 2.6 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.6 | 9.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.6 | 1.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 14.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.5 | 2.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 1.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 2.4 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 21.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 2.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 10.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 6.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 6.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 6.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 2.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 4.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 2.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 2.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 1.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 1.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 10.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 18.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 0.1 | 2.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.7 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 1.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 4.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 3.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 3.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 8.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 13.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 7.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 7.9 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 2.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 4.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 2.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 3.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 6.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.0 | GO:0030594 | neurotransmitter receptor activity(GO:0030594) |
| 0.0 | 25.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 4.7 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 8.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 7.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 7.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 16.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.9 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 10.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 2.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 7.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 13.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 6.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 3.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.3 | 4.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 6.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 7.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 2.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 2.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |