Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
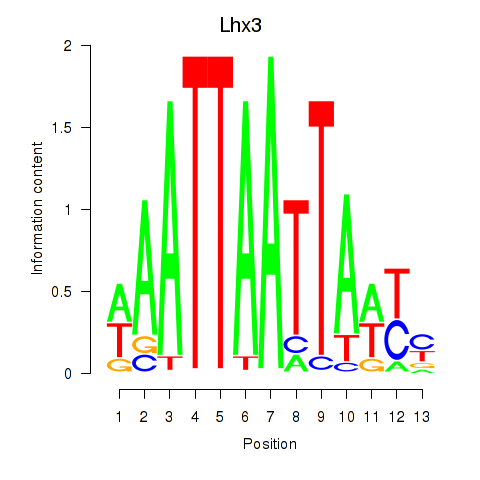
Results for Lhx3
Z-value: 0.53
Transcription factors associated with Lhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx3
|
ENSRNOG00000018427 | LIM homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx3 | rn6_v1_chr3_-_3661810_3661810 | 0.09 | 9.4e-02 | Click! |
Activity profile of Lhx3 motif
Sorted Z-values of Lhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_71139267 | 46.40 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr3_+_159368273 | 23.84 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr18_+_59748444 | 20.35 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr10_+_103206014 | 17.89 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr11_+_36851038 | 15.57 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr6_+_64789940 | 14.17 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr15_-_93307420 | 13.89 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr2_+_54466280 | 13.48 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr13_+_47572219 | 13.03 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr8_-_7426611 | 11.95 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr9_+_73418607 | 11.95 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr9_+_73378057 | 11.17 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr5_+_124442293 | 11.07 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr14_+_39964588 | 10.81 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr7_-_105592804 | 10.76 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr1_+_238222521 | 10.70 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr13_+_84474319 | 10.61 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr20_+_34258791 | 10.27 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr6_+_48452369 | 9.23 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr20_-_4542073 | 8.61 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr14_-_84334066 | 8.23 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chr4_-_55011415 | 7.92 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr6_+_2216623 | 7.89 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrX_+_151103576 | 7.55 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr3_-_67787990 | 7.04 |
ENSRNOT00000064851
|
Nckap1
|
NCK-associated protein 1 |
| chr13_-_76049363 | 6.45 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr16_+_23447366 | 6.36 |
ENSRNOT00000068629
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_-_181900856 | 6.13 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr15_+_59678165 | 5.81 |
ENSRNOT00000074868
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr18_+_16146447 | 5.81 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr17_-_89923423 | 5.67 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chrX_+_20520034 | 5.39 |
ENSRNOT00000093170
|
FAM120C
|
family with sequence similarity 120C |
| chr12_-_2174131 | 4.92 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr12_+_41486076 | 4.32 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr10_+_53778662 | 4.31 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr1_-_264975132 | 4.21 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr9_-_88086488 | 4.06 |
ENSRNOT00000019579
|
Irs1
|
insulin receptor substrate 1 |
| chr7_+_44009069 | 4.06 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chrX_-_29648359 | 3.87 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr11_-_68989988 | 3.61 |
ENSRNOT00000003013
|
Hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr1_+_185863043 | 3.42 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr5_+_6373583 | 3.11 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr14_-_21909646 | 3.05 |
ENSRNOT00000088024
|
Csn1s2b
|
casein alpha s2-like B |
| chr12_-_46493203 | 3.01 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr6_-_142418779 | 3.00 |
ENSRNOT00000072280
ENSRNOT00000065808 |
AABR07065814.1
|
|
| chr18_+_29993361 | 2.99 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr10_-_34301197 | 2.97 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr18_-_43945273 | 2.89 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chrX_-_111191932 | 2.86 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr20_-_13994794 | 2.84 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr2_+_30685840 | 2.80 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr8_-_43524220 | 2.63 |
ENSRNOT00000088755
|
Olr1315
|
olfactory receptor 1315 |
| chrM_+_9870 | 2.46 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chrX_+_14019961 | 2.44 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr2_-_233743866 | 2.34 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr11_-_11585078 | 2.26 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chrX_-_124252447 | 2.23 |
ENSRNOT00000061546
|
Rhox12
|
reproductive homeobox 12 |
| chr6_+_18880737 | 2.10 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_-_149444548 | 2.05 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr1_+_79631668 | 2.02 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr15_-_46166335 | 1.93 |
ENSRNOT00000059215
|
Defb42
|
defensin beta 42 |
| chr14_+_7026769 | 1.89 |
ENSRNOT00000071955
|
LOC100359907
|
secretory calcium-binding phosphoprotein proline-glutamine rich 1-like |
| chr2_+_145174876 | 1.63 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr3_+_119561290 | 1.59 |
ENSRNOT00000015843
|
Blvra
|
biliverdin reductase A |
| chr2_-_35550204 | 1.57 |
ENSRNOT00000084377
|
Olr1280
|
olfactory receptor 1280 |
| chr10_+_67810810 | 1.56 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr8_-_44565861 | 1.48 |
ENSRNOT00000049921
|
AABR07069997.1
|
|
| chr15_+_27438853 | 1.37 |
ENSRNOT00000011636
|
Olr1622
|
olfactory receptor 1622 |
| chr2_+_226563050 | 1.36 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_813517 | 1.24 |
ENSRNOT00000041332
|
Vom2r5
|
vomeronasal 2 receptor, 5 |
| chr16_-_75241303 | 1.24 |
ENSRNOT00000058056
|
Defb2
|
defensin beta 2 |
| chr9_-_121725716 | 1.02 |
ENSRNOT00000087405
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr8_+_19888667 | 0.93 |
ENSRNOT00000078593
|
Zfp317
|
zinc finger protein 317 |
| chr14_-_1785698 | 0.92 |
ENSRNOT00000071182
|
Vom2r73
|
vomeronasal 2 receptor, 73 |
| chr1_+_230628996 | 0.90 |
ENSRNOT00000017604
|
Olr376
|
olfactory receptor 376 |
| chr18_-_36322320 | 0.82 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
| chr1_+_22319353 | 0.80 |
ENSRNOT00000038523
|
Taar9
|
trace amine-associated receptor 9 |
| chr14_-_24123253 | 0.80 |
ENSRNOT00000002743
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chr15_-_54528480 | 0.76 |
ENSRNOT00000066888
|
Fndc3a
|
fibronectin type III domain containing 3a |
| chrM_+_9451 | 0.74 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_-_69016426 | 0.73 |
ENSRNOT00000093436
|
Vom2r3
|
|
| chr10_+_67810655 | 0.73 |
ENSRNOT00000064285
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr2_+_239415046 | 0.63 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chrX_+_112311251 | 0.63 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr14_+_23405717 | 0.60 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr2_-_198706428 | 0.60 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr11_-_43594659 | 0.58 |
ENSRNOT00000086976
|
Olr1555
|
olfactory receptor 1555 |
| chr3_+_77289805 | 0.57 |
ENSRNOT00000040581
|
LOC100910858
|
olfactory receptor 4P4-like |
| chr8_+_117297670 | 0.51 |
ENSRNOT00000082628
|
Qars
|
glutaminyl-tRNA synthetase |
| chrX_-_120521871 | 0.50 |
ENSRNOT00000080863
|
LOC100362376
|
LRRGT00025-like |
| chr14_+_36047144 | 0.39 |
ENSRNOT00000003088
|
Lnx1
|
ligand of numb-protein X 1 |
| chr3_+_76052230 | 0.16 |
ENSRNOT00000039963
|
LOC100911380
|
olfactory receptor 5W2-like |
| chr15_+_35889080 | 0.15 |
ENSRNOT00000075355
|
LOC100910640
|
olfactory receptor 144-like |
| chr10_+_112056994 | 0.11 |
ENSRNOT00000073032
|
Vom2r65
|
vomeronasal 2 receptor, 65 |
| chr10_-_60032218 | 0.11 |
ENSRNOT00000072328
|
Olr1481
|
olfactory receptor 1481 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 4.5 | 13.5 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 4.3 | 13.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 4.0 | 12.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 3.9 | 15.6 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 1.6 | 14.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.4 | 5.7 | GO:0030242 | pexophagy(GO:0030242) |
| 1.4 | 7.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 1.3 | 10.7 | GO:0042905 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 5.8 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.9 | 8.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.8 | 4.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.8 | 3.9 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 6.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.7 | 7.9 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.7 | 2.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.7 | 3.4 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.6 | 4.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.6 | 23.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.5 | 2.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.5 | 2.3 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.5 | 2.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.4 | 4.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 2.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 4.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.6 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.3 | 6.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 10.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 1.0 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.2 | 3.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 20.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 10.8 | GO:0007616 | long-term memory(GO:0007616) |
| 0.2 | 10.6 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 2.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 23.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.2 | 11.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 16.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 8.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 3.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 2.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 6.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.7 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 1.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 2.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 2.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 2.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 3.0 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.5 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 23.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.9 | 13.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.4 | 4.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.0 | 15.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 7.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 14.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.5 | 28.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 4.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 20.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 11.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 2.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 13.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 4.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 6.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 8.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 5.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 7.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 8.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 7.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 3.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 9.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 8.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 15.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.2 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 3.6 | 10.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 2.0 | 7.9 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.3 | 23.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.2 | 9.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.0 | 9.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.0 | 23.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.9 | 10.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.8 | 4.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.7 | 3.6 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.6 | 10.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 5.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.5 | 2.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 4.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 2.8 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.5 | 6.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 3.0 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.3 | 2.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 6.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 5.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 2.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 16.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 7.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 3.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 16.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 11.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 3.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 2.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 8.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.1 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 4.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 4.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.4 | 23.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 4.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 6.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 16.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 7.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 24.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 1.3 | 10.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 22.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 14.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 7.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 4.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 4.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 6.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 5.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |