Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
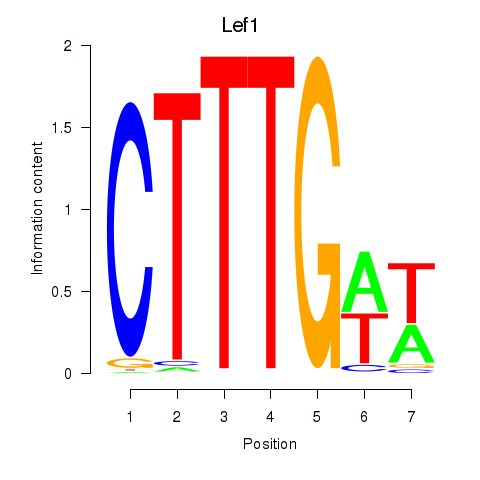
Results for Lef1
Z-value: 0.72
Transcription factors associated with Lef1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lef1
|
ENSRNOG00000010121 | lymphoid enhancer binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lef1 | rn6_v1_chr2_+_236233239_236233239 | -0.07 | 2.3e-01 | Click! |
Activity profile of Lef1 motif
Sorted Z-values of Lef1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_51643140 | 28.77 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr7_-_98098268 | 23.74 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr16_-_39476025 | 16.51 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr9_-_85243001 | 15.44 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr8_+_49418965 | 15.40 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chr8_-_14880644 | 15.39 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr14_+_66598259 | 15.16 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr10_-_27179254 | 14.69 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr3_+_131351587 | 14.38 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr11_+_9642365 | 14.21 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr4_+_142453013 | 14.20 |
ENSRNOT00000056573
|
AABR07061755.1
|
|
| chr19_-_19509087 | 13.61 |
ENSRNOT00000019301
|
Nkd1
|
naked cuticle homolog 1 |
| chr10_-_27179900 | 13.34 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr4_+_172942020 | 12.30 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chrX_-_32095867 | 12.12 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr13_-_91872954 | 11.99 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr13_-_72063347 | 11.88 |
ENSRNOT00000090544
ENSRNOT00000003869 |
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr1_+_59156251 | 11.63 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr8_+_128824508 | 10.93 |
ENSRNOT00000025343
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr5_-_7874909 | 10.57 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr8_-_84506328 | 10.48 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr17_+_47721977 | 10.47 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr1_+_151439409 | 10.34 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chr2_-_220535751 | 10.17 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chrX_-_68562301 | 10.10 |
ENSRNOT00000076720
|
Ophn1
|
oligophrenin 1 |
| chr3_+_154395187 | 10.08 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr4_+_99618622 | 10.04 |
ENSRNOT00000064772
|
Reep1
|
receptor accessory protein 1 |
| chr15_+_52767442 | 9.94 |
ENSRNOT00000014441
|
LOC108348161
|
phytanoyl-CoA hydroxylase-interacting protein |
| chr5_+_104362971 | 9.93 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chrX_+_156655960 | 9.61 |
ENSRNOT00000085723
|
Mecp2
|
methyl CpG binding protein 2 |
| chr13_-_107831014 | 9.41 |
ENSRNOT00000003684
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr6_-_147172022 | 9.18 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr8_+_79606789 | 9.14 |
ENSRNOT00000087114
|
Pygo1
|
pygopus family PHD finger 1 |
| chr6_+_147876237 | 9.05 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr2_-_212852870 | 9.00 |
ENSRNOT00000066218
|
Ntng1
|
netrin G1 |
| chr20_-_27208041 | 8.76 |
ENSRNOT00000084720
|
Rufy2
|
RUN and FYVE domain containing 2 |
| chr2_+_144861455 | 8.72 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr15_+_52148379 | 8.69 |
ENSRNOT00000074912
|
Phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr5_-_34813116 | 8.68 |
ENSRNOT00000017479
|
Nkain3
|
Sodium/potassium transporting ATPase interacting 3 |
| chr6_-_147172813 | 8.55 |
ENSRNOT00000066545
|
Itgb8
|
integrin subunit beta 8 |
| chr8_-_73164620 | 8.33 |
ENSRNOT00000031988
|
Tln2
|
talin 2 |
| chr2_-_57935334 | 8.30 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr8_+_49676540 | 8.27 |
ENSRNOT00000022032
ENSRNOT00000082205 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr2_-_147693082 | 8.16 |
ENSRNOT00000022507
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr6_+_147876557 | 8.15 |
ENSRNOT00000080090
|
Tmem196
|
transmembrane protein 196 |
| chr7_-_93826665 | 7.84 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr5_-_168734296 | 7.81 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr4_-_82209933 | 7.66 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr17_+_13670520 | 7.66 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr4_-_82295470 | 7.64 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr3_-_107760550 | 7.56 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr9_+_2202511 | 7.54 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr15_-_41338284 | 7.38 |
ENSRNOT00000077225
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr10_+_98706960 | 7.26 |
ENSRNOT00000006217
|
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr10_+_81913689 | 7.03 |
ENSRNOT00000003780
|
Tob1
|
transducer of ErbB-2.1 |
| chr9_+_66335492 | 6.93 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr2_-_140618405 | 6.85 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr12_+_27155587 | 6.84 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr3_-_46726946 | 6.73 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr13_-_111581018 | 6.62 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr3_-_168018410 | 6.50 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chrX_+_9436707 | 6.28 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr9_-_61810417 | 6.18 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr20_+_48335540 | 6.17 |
ENSRNOT00000000352
|
Cd24
|
CD24 molecule |
| chr15_+_86243148 | 6.16 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr14_+_63095720 | 6.12 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr2_+_179952227 | 6.05 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr5_+_4373626 | 5.97 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr13_+_106463368 | 5.84 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chr13_+_71110064 | 5.75 |
ENSRNOT00000088329
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr14_-_87701884 | 5.74 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr18_+_30909490 | 5.61 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr2_-_256154584 | 5.57 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr13_-_81214494 | 5.50 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr17_-_389967 | 5.49 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chrX_-_159891326 | 5.44 |
ENSRNOT00000001154
|
Rbmx
|
RNA binding motif protein, X-linked |
| chr7_+_144628120 | 5.36 |
ENSRNOT00000022247
|
Hoxc5
|
homeo box C5 |
| chr2_+_188528979 | 5.35 |
ENSRNOT00000087934
|
Thbs3
|
thrombospondin 3 |
| chrX_-_18726732 | 5.23 |
ENSRNOT00000026214
|
Ubqln2
|
ubiquilin 2 |
| chr11_-_62451149 | 4.97 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_82203154 | 4.93 |
ENSRNOT00000086210
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr7_-_107009330 | 4.91 |
ENSRNOT00000074573
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr2_+_196013799 | 4.91 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr10_+_84119884 | 4.69 |
ENSRNOT00000009951
|
Hoxb9
|
homeo box B9 |
| chr9_+_40817654 | 4.62 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr18_+_14756684 | 4.45 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr6_-_27721120 | 4.38 |
ENSRNOT00000071425
|
Rab10
|
RAB10, member RAS oncogene family |
| chr18_+_30900291 | 4.35 |
ENSRNOT00000060461
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr6_+_28515025 | 4.33 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr8_+_111210811 | 4.26 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr17_-_84830185 | 4.25 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chr13_+_5480150 | 4.18 |
ENSRNOT00000057873
|
AABR07019870.1
|
|
| chr1_+_41590494 | 4.14 |
ENSRNOT00000089984
|
Esr1
|
estrogen receptor 1 |
| chr15_-_2966576 | 3.94 |
ENSRNOT00000070893
ENSRNOT00000017383 |
Kat6b
|
lysine acetyltransferase 6B |
| chr1_-_263431290 | 3.94 |
ENSRNOT00000022633
|
Slc25a28
|
solute carrier family 25 member 28 |
| chr3_+_116899878 | 3.89 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr16_+_37500017 | 3.80 |
ENSRNOT00000081634
|
AC135696.1
|
high mobility group box 1 (Hmgb1), mRNA |
| chr4_-_82215022 | 3.63 |
ENSRNOT00000010256
|
Hoxa11
|
homeobox A11 |
| chr14_-_37770059 | 3.59 |
ENSRNOT00000029721
|
Slc10a4
|
solute carrier family 10, member 4 |
| chr4_+_78240385 | 3.57 |
ENSRNOT00000011041
|
Zfp775
|
zinc finger protein 775 |
| chr3_+_58164931 | 3.54 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr3_+_151691515 | 3.48 |
ENSRNOT00000052236
|
AC118414.1
|
|
| chr16_-_73152921 | 3.46 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chrX_-_134866210 | 3.42 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr2_+_150756185 | 3.36 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr6_+_73358112 | 3.35 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr7_-_36408588 | 3.34 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chrX_+_86126157 | 3.33 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr3_-_43119159 | 3.27 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr15_-_52214616 | 3.23 |
ENSRNOT00000015035
|
Sftpc
|
surfactant protein C |
| chr10_-_43253296 | 3.15 |
ENSRNOT00000003486
|
Hand1
|
heart and neural crest derivatives expressed 1 |
| chr10_+_75365822 | 3.13 |
ENSRNOT00000055705
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr10_-_89700283 | 3.12 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr10_-_86004096 | 3.12 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr1_-_248898607 | 3.03 |
ENSRNOT00000074393
|
NEWGENE_1307313
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr13_-_111474411 | 2.98 |
ENSRNOT00000072729
|
Hhat
|
hedgehog acyltransferase |
| chr2_-_250241590 | 2.84 |
ENSRNOT00000077221
ENSRNOT00000067502 |
Lmo4
|
LIM domain only 4 |
| chr4_+_44321883 | 2.69 |
ENSRNOT00000091095
|
Tes
|
testin LIM domain protein |
| chr9_-_38196273 | 2.69 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr7_-_132984110 | 2.67 |
ENSRNOT00000046626
|
Hmgb1-ps3
|
high mobility group box 1, pseudogene 3 |
| chr18_-_15688284 | 2.66 |
ENSRNOT00000091816
|
Dsg3
|
desmoglein 3 |
| chr3_+_149741312 | 2.63 |
ENSRNOT00000070854
|
LOC100911637
|
high mobility group protein B1-like |
| chr1_-_253185533 | 2.57 |
ENSRNOT00000067822
|
Pank1
|
pantothenate kinase 1 |
| chr8_+_117620317 | 2.53 |
ENSRNOT00000084220
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr2_-_143104412 | 2.42 |
ENSRNOT00000058116
|
Ufm1
|
ubiquitin-fold modifier 1 |
| chr10_+_10644572 | 2.25 |
ENSRNOT00000004026
|
Ppl
|
periplakin |
| chr1_-_24302298 | 2.19 |
ENSRNOT00000083452
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr13_-_52514875 | 2.17 |
ENSRNOT00000064758
|
Nav1
|
neuron navigator 1 |
| chr9_-_4447715 | 2.16 |
ENSRNOT00000061882
|
Kat2b
|
lysine acetyltransferase 2B |
| chr2_-_211017778 | 2.11 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr6_+_99625306 | 2.06 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr1_-_198316882 | 2.06 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr10_+_84182118 | 2.06 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr4_+_179481263 | 2.02 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr4_+_82300778 | 1.98 |
ENSRNOT00000075254
|
Hoxa11-as
|
homeobox A11, opposite strand |
| chr2_+_142686724 | 1.91 |
ENSRNOT00000014614
|
Proser1
|
proline and serine rich 1 |
| chr16_-_74264142 | 1.73 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr3_+_148541909 | 1.71 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr2_+_51672722 | 1.67 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr5_-_169017295 | 1.61 |
ENSRNOT00000067481
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr9_-_3697736 | 1.61 |
ENSRNOT00000087331
|
Kcnh8
|
potassium voltage-gated channel subfamily H member 8 |
| chr3_-_9262628 | 1.60 |
ENSRNOT00000013286
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr7_+_24939498 | 1.58 |
ENSRNOT00000010601
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr8_+_63600663 | 1.58 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr4_-_60548021 | 1.54 |
ENSRNOT00000065546
|
AABR07060165.1
|
|
| chr4_-_82300503 | 1.53 |
ENSRNOT00000071568
|
Hoxa11
|
homeobox A11 |
| chr8_-_118182559 | 1.52 |
ENSRNOT00000084838
|
Dhx30
|
DExH-box helicase 30 |
| chr2_+_204427608 | 1.49 |
ENSRNOT00000083374
|
Nhlh2
|
nescient helix loop helix 2 |
| chr2_-_211001258 | 1.49 |
ENSRNOT00000037336
|
Atxn7l2
|
ataxin 7-like 2 |
| chr12_-_47793534 | 1.45 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr6_-_55524436 | 1.43 |
ENSRNOT00000006752
|
Tspan13
|
tetraspanin 13 |
| chr4_+_82214342 | 1.41 |
ENSRNOT00000066360
|
Hoxa11-as
|
homeobox A11, opposite strand |
| chrX_-_115175299 | 1.40 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr10_+_64398339 | 1.38 |
ENSRNOT00000056278
|
Mrm3
|
mitochondrial rRNA methyltransferase 3 |
| chr2_+_205525204 | 1.34 |
ENSRNOT00000091210
|
Csde1
|
cold shock domain containing E1 |
| chr20_+_42966140 | 1.25 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr10_+_94407559 | 1.21 |
ENSRNOT00000013046
|
Ddx42
|
DEAD-box helicase 42 |
| chr8_+_70630767 | 1.10 |
ENSRNOT00000051353
|
Igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr1_-_218100272 | 0.91 |
ENSRNOT00000028411
ENSRNOT00000088588 |
Ccnd1
|
cyclin D1 |
| chr10_-_64398294 | 0.88 |
ENSRNOT00000010386
|
Glod4
|
glyoxalase domain containing 4 |
| chr19_-_25914689 | 0.87 |
ENSRNOT00000091994
ENSRNOT00000039354 |
Nf1x
|
nuclear factor 1 X |
| chr4_+_157554794 | 0.86 |
ENSRNOT00000024116
|
Ing4
|
inhibitor of growth family, member 4 |
| chr13_-_82006005 | 0.83 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr13_-_84619023 | 0.82 |
ENSRNOT00000089827
ENSRNOT00000045056 |
Pogk
|
pogo transposable element with KRAB domain |
| chr8_-_78397123 | 0.82 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr5_+_122100099 | 0.70 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr10_-_83128297 | 0.68 |
ENSRNOT00000082160
|
Kat7
|
lysine acetyltransferase 7 |
| chr11_-_35697072 | 0.67 |
ENSRNOT00000039999
|
Erg
|
ERG, ETS transcription factor |
| chr15_+_105851542 | 0.62 |
ENSRNOT00000086959
|
Rap2a
|
RAS related protein 2a |
| chr13_+_89797800 | 0.61 |
ENSRNOT00000005811
|
Usf1
|
upstream transcription factor 1 |
| chr9_+_37593032 | 0.61 |
ENSRNOT00000051890
|
AABR07067274.1
|
|
| chrX_-_26376467 | 0.55 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr7_-_50034932 | 0.55 |
ENSRNOT00000081885
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr9_+_46657922 | 0.52 |
ENSRNOT00000019180
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_-_59514939 | 0.49 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr15_+_37171052 | 0.48 |
ENSRNOT00000011684
|
Zmym2
|
zinc finger MYM-type containing 2 |
| chr9_-_111213098 | 0.44 |
ENSRNOT00000087355
|
Gin1
|
gypsy retrotransposon integrase 1 |
| chr6_-_99843245 | 0.42 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr7_+_58570579 | 0.39 |
ENSRNOT00000081227
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr3_+_47677720 | 0.36 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr1_-_167971151 | 0.31 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr5_-_144479306 | 0.30 |
ENSRNOT00000087697
|
Ago1
|
argonaute 1, RISC catalytic component |
| chr6_+_8669722 | 0.29 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr8_-_129371973 | 0.27 |
ENSRNOT00000042614
|
Snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr5_-_139748489 | 0.26 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr10_-_85574889 | 0.20 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr2_-_139528162 | 0.18 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr16_-_71203609 | 0.18 |
ENSRNOT00000088458
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr16_+_49185560 | 0.07 |
ENSRNOT00000014360
|
Helt
|
helt bHLH transcription factor |
| chr10_-_89699836 | 0.06 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr20_-_31598118 | 0.04 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr6_+_44009872 | 0.00 |
ENSRNOT00000082657
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lef1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 29.9 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 4.7 | 14.2 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 4.4 | 44.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 4.0 | 12.1 | GO:1903598 | angiotensin-mediated drinking behavior(GO:0003051) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 3.4 | 10.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 3.2 | 9.6 | GO:2000820 | positive regulation of histone H3-K9 trimethylation(GO:1900114) negative regulation of dendrite extension(GO:1903860) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 3.1 | 9.4 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 2.7 | 13.6 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 2.6 | 7.8 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 2.1 | 8.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.1 | 6.2 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 2.0 | 28.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.8 | 7.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.7 | 5.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 1.5 | 12.3 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 1.5 | 11.9 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.4 | 5.5 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.3 | 6.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 1.2 | 3.5 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 1.1 | 15.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.1 | 4.4 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 1.0 | 4.1 | GO:1990375 | baculum development(GO:1990375) |
| 1.0 | 6.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.0 | 8.2 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 1.0 | 6.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.0 | 17.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.9 | 3.4 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.8 | 3.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.8 | 8.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 | 2.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.7 | 10.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.7 | 7.0 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA catabolic process(GO:1902373) |
| 0.7 | 6.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.7 | 3.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.6 | 3.2 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.6 | 2.5 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.6 | 15.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.6 | 1.7 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.6 | 1.7 | GO:0071338 | mammary gland bud morphogenesis(GO:0060648) submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.5 | 5.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.5 | 7.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.5 | 3.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.5 | 2.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.5 | 5.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.5 | 2.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.5 | 6.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.5 | 10.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.4 | 5.8 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.4 | 9.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.4 | 12.8 | GO:0060065 | uterus development(GO:0060065) |
| 0.4 | 16.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 6.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.4 | 4.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.4 | 1.5 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.4 | 11.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 8.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 1.6 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.3 | 10.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.3 | 0.8 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.2 | 1.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 1.7 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.2 | 4.9 | GO:0061548 | ganglion development(GO:0061548) |
| 0.2 | 2.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 1.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 0.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 0.7 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.2 | 1.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 10.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.2 | 3.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 0.6 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) positive regulation of transcription by glucose(GO:0046016) |
| 0.2 | 10.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 2.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 0.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 5.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 8.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 3.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 2.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.1 | 3.9 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 3.9 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 10.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 7.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 4.3 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 3.3 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 5.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 12.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 0.9 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 2.8 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 0.6 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 3.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 5.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 12.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.3 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.1 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 3.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 7.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 1.5 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 3.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 2.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 7.2 | GO:0007611 | learning or memory(GO:0007611) |
| 0.0 | 5.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 4.8 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 3.5 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 2.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 1.4 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.4 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.7 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 2.2 | 44.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.2 | 28.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.2 | 9.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.1 | 5.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.0 | 15.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 10.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.5 | 2.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.5 | 38.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.5 | 1.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.5 | 10.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.5 | 13.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 6.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.4 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 4.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.4 | 3.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 7.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.4 | 6.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 16.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 4.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 7.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 1.2 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.3 | 9.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 11.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 24.6 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 3.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 7.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 6.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 4.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 15.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 5.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 20.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 28.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 2.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 15.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 3.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 20.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 10.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 5.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 7.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 2.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 6.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 8.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 16.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.6 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 3.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 2.6 | 10.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 2.4 | 11.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 2.2 | 17.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.0 | 10.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 2.0 | 28.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.9 | 15.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.9 | 9.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.9 | 9.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.7 | 8.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.6 | 30.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 1.5 | 5.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.2 | 14.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.1 | 3.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.9 | 10.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.8 | 4.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.6 | 2.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.6 | 17.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 7.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.6 | 3.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 10.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 6.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 3.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.4 | 7.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 2.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 2.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 6.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 3.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 6.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 0.8 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.3 | 6.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 15.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 8.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 10.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 1.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 14.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 4.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 3.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 5.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 4.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 16.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 6.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 3.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 13.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 2.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 3.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 6.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 4.6 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 7.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.0 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 4.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 9.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.6 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 18.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 14.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 10.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 12.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 24.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 8.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 15.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 14.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 6.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 12.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 8.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 14.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 9.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 7.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 7.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 3.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 8.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 9.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 28.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 9.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.6 | 14.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.6 | 20.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 7.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 12.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 6.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 7.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 22.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 3.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 6.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 2.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 13.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 2.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 8.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 5.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 7.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 6.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 3.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 4.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |