Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
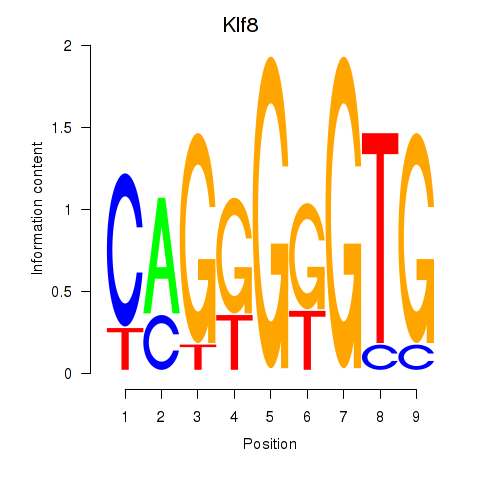
Results for Klf8
Z-value: 0.40
Transcription factors associated with Klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf8
|
ENSRNOG00000039057 | Kruppel-like factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf8 | rn6_v1_chrX_-_18890090_18890090 | -0.08 | 1.7e-01 | Click! |
Activity profile of Klf8 motif
Sorted Z-values of Klf8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_15273933 | 11.48 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr3_-_160730360 | 10.15 |
ENSRNOT00000075864
|
RGD1563818
|
similar to secretory leukocyte protease inhibitor |
| chrX_-_38196060 | 8.30 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr1_-_53087474 | 7.19 |
ENSRNOT00000017302
|
Ccr6
|
C-C motif chemokine receptor 6 |
| chr7_+_145068286 | 6.49 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr19_-_37970537 | 5.85 |
ENSRNOT00000034722
|
Dpep2
|
dipeptidase 2 |
| chr10_-_90999506 | 5.67 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr4_+_161685258 | 5.16 |
ENSRNOT00000008012
ENSRNOT00000008003 |
Foxm1
|
forkhead box M1 |
| chr1_-_198460126 | 4.92 |
ENSRNOT00000082940
ENSRNOT00000086019 |
Maz
|
MYC associated zinc finger protein |
| chr1_-_89488223 | 4.87 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr2_+_205160405 | 4.80 |
ENSRNOT00000035605
|
Tspan2
|
tetraspanin 2 |
| chr6_-_142372031 | 4.40 |
ENSRNOT00000063929
|
AABR07065814.3
|
|
| chr14_-_2700977 | 4.20 |
ENSRNOT00000000086
|
Mtf2
|
metal response element binding transcription factor 2 |
| chr1_-_42467586 | 4.10 |
ENSRNOT00000029416
|
Fbxo5
|
F-box protein 5 |
| chr10_-_15590220 | 4.06 |
ENSRNOT00000048977
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr1_+_221872220 | 4.04 |
ENSRNOT00000028651
|
Nrxn2
|
neurexin 2 |
| chr15_+_31243097 | 4.03 |
ENSRNOT00000042721
|
LOC100360891
|
RGD1359684 protein-like |
| chr7_-_121311565 | 4.01 |
ENSRNOT00000092252
|
Rpl3
|
ribosomal protein L3 |
| chr8_-_67232298 | 3.94 |
ENSRNOT00000049441
|
Spesp1
|
sperm equatorial segment protein 1 |
| chr1_+_63964155 | 3.76 |
ENSRNOT00000078512
|
AABR07002001.1
|
|
| chr12_+_23544287 | 3.63 |
ENSRNOT00000001938
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr4_-_45332420 | 3.43 |
ENSRNOT00000083039
|
Wnt2
|
wingless-type MMTV integration site family member 2 |
| chr7_+_141249044 | 3.17 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr9_+_100137881 | 3.14 |
ENSRNOT00000036328
|
Gpr35
|
G protein-coupled receptor 35 |
| chr10_-_89454681 | 3.08 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr14_-_78377825 | 2.93 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr6_-_146195819 | 2.75 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr4_-_100883275 | 2.64 |
ENSRNOT00000022846
|
LOC100364435
|
thymosin, beta 10-like |
| chr1_-_167347490 | 2.60 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr1_-_73682247 | 2.47 |
ENSRNOT00000079498
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr12_-_22478752 | 2.42 |
ENSRNOT00000089392
ENSRNOT00000086915 |
Ache
|
acetylcholinesterase |
| chr17_-_32558180 | 2.33 |
ENSRNOT00000022681
|
Serpinb1b
|
serine (or cysteine) peptidase inhibitor, clade B, member 1b |
| chr7_+_38945836 | 2.22 |
ENSRNOT00000006455
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr20_-_4943564 | 2.21 |
ENSRNOT00000049310
|
RT1-CE1
|
RT1 class I, locus1 |
| chr10_+_55169282 | 2.17 |
ENSRNOT00000005423
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr11_-_87236445 | 2.08 |
ENSRNOT00000057806
|
Tssk2
|
testis-specific serine kinase 2 |
| chr1_-_84168044 | 2.03 |
ENSRNOT00000084586
ENSRNOT00000028348 |
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr1_-_142183884 | 1.95 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr15_+_34431357 | 1.93 |
ENSRNOT00000027604
|
Nop9
|
NOP9 nucleolar protein |
| chr12_+_13716596 | 1.90 |
ENSRNOT00000080216
|
Actb
|
actin, beta |
| chr1_-_84812486 | 1.87 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr13_+_79378733 | 1.76 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr1_+_99648658 | 1.75 |
ENSRNOT00000039531
|
Klk13
|
kallikrein related-peptidase 13 |
| chr1_+_72636959 | 1.73 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr3_+_152909189 | 1.72 |
ENSRNOT00000066341
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr12_-_47588335 | 1.71 |
ENSRNOT00000001595
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr4_+_10823281 | 1.68 |
ENSRNOT00000066739
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr3_-_161917285 | 1.64 |
ENSRNOT00000025150
|
Cdh22
|
cadherin 22 |
| chr15_+_24267323 | 1.63 |
ENSRNOT00000015778
|
Fbxo34
|
F-box protein 34 |
| chr2_+_262914327 | 1.61 |
ENSRNOT00000029312
|
Negr1
|
neuronal growth regulator 1 |
| chr7_-_144960527 | 1.60 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr1_-_167347662 | 1.60 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr18_+_28594892 | 1.57 |
ENSRNOT00000067702
|
Ube2d2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr18_+_48132414 | 1.57 |
ENSRNOT00000050631
|
Snx2
|
sorting nexin 2 |
| chr5_-_139933764 | 1.57 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr19_+_38669230 | 1.56 |
ENSRNOT00000027273
|
Cdh3
|
cadherin 3 |
| chr4_-_58195143 | 1.51 |
ENSRNOT00000033706
|
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr14_-_83062302 | 1.49 |
ENSRNOT00000086769
ENSRNOT00000085735 |
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr20_-_3728844 | 1.43 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr2_-_252359798 | 1.40 |
ENSRNOT00000021063
|
Spata1
|
spermatogenesis associated 1 |
| chr3_-_160777092 | 1.40 |
ENSRNOT00000076624
|
Slpil3
|
antileukoproteinase-like 3 |
| chr6_-_76079283 | 1.37 |
ENSRNOT00000082198
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr9_+_14551758 | 1.37 |
ENSRNOT00000017157
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr17_+_36334589 | 1.30 |
ENSRNOT00000081368
|
E2f3
|
E2F transcription factor 3 |
| chr10_-_109827143 | 1.30 |
ENSRNOT00000054948
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr1_-_221041401 | 1.24 |
ENSRNOT00000064136
|
Pcnx3
|
pecanex homolog 3 (Drosophila) |
| chr4_-_115332052 | 1.18 |
ENSRNOT00000017643
|
Clec4f
|
C-type lectin domain family 4, member F |
| chr1_+_99781237 | 1.17 |
ENSRNOT00000025489
|
Klk5
|
kallikrein related-peptidase 5 |
| chr3_-_72113680 | 1.12 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chr10_-_29450644 | 1.12 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr10_-_13075864 | 1.10 |
ENSRNOT00000005220
|
Paqr4
|
progestin and adipoQ receptor family member 4 |
| chr10_+_69434941 | 1.09 |
ENSRNOT00000009756
|
Ccl11
|
C-C motif chemokine ligand 11 |
| chr3_+_113976687 | 1.08 |
ENSRNOT00000022437
|
Eif3j
|
eukaryotic translation initiation factor 3, subunit J |
| chr7_+_142877086 | 1.05 |
ENSRNOT00000088730
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chr2_+_24024791 | 1.05 |
ENSRNOT00000089599
ENSRNOT00000066302 |
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr3_-_5693147 | 1.04 |
ENSRNOT00000008660
|
Fam163b
|
family with sequence similarity 163, member B |
| chr10_+_56237755 | 1.03 |
ENSRNOT00000016163
|
Fxr2
|
FMR1 autosomal homolog 2 |
| chr2_+_251817694 | 1.03 |
ENSRNOT00000019964
|
RGD1560065
|
similar to RIKEN cDNA 2410004B18 |
| chr1_-_80271001 | 0.97 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr6_+_26517840 | 0.97 |
ENSRNOT00000031842
|
Ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr1_+_192025710 | 0.97 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr1_-_72461547 | 0.95 |
ENSRNOT00000022480
|
Ssc5d
|
scavenger receptor cysteine rich family member with 5 domains |
| chr1_+_225015616 | 0.93 |
ENSRNOT00000026416
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr6_-_142178771 | 0.91 |
ENSRNOT00000071577
|
Ighv12-3
|
immunoglobulin heavy variable V12-3 |
| chr6_-_141866756 | 0.90 |
ENSRNOT00000068561
|
AABR07065804.1
|
|
| chr6_-_76079664 | 0.90 |
ENSRNOT00000033533
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr2_+_231962517 | 0.88 |
ENSRNOT00000014570
|
Neurog2
|
neurogenin 2 |
| chr2_+_17616401 | 0.82 |
ENSRNOT00000064991
|
Edil3
|
EGF like repeats and discoidin domains 3 |
| chr1_-_73753128 | 0.82 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr8_-_123371257 | 0.81 |
ENSRNOT00000017243
|
Stt3b
|
STT3B, catalytic subunit of the oligosaccharyltransferase complex |
| chr19_-_11669578 | 0.76 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr8_+_43348973 | 0.75 |
ENSRNOT00000007862
|
Olr1308
|
olfactory receptor 1308 |
| chr8_-_117797427 | 0.74 |
ENSRNOT00000033719
|
Trex1
|
three prime repair exonuclease 1 |
| chr1_+_214664537 | 0.73 |
ENSRNOT00000075002
|
AABR07006030.1
|
|
| chr2_-_84437084 | 0.72 |
ENSRNOT00000077598
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr10_+_45798797 | 0.70 |
ENSRNOT00000039012
|
Jmjd4
|
jumonji domain containing 4 |
| chr11_+_34598492 | 0.70 |
ENSRNOT00000065600
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr8_+_32018560 | 0.69 |
ENSRNOT00000007358
|
Adamts8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr3_+_113131327 | 0.60 |
ENSRNOT00000018460
|
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr5_+_173256637 | 0.59 |
ENSRNOT00000025531
|
Ccnl2
|
cyclin L2 |
| chr18_+_3861539 | 0.58 |
ENSRNOT00000015363
|
Lama3
|
laminin subunit alpha 3 |
| chr20_+_4576514 | 0.58 |
ENSRNOT00000090125
ENSRNOT00000047370 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr1_-_4653210 | 0.57 |
ENSRNOT00000019121
|
Rab32
|
RAB32, member RAS oncogene family |
| chr1_+_261337594 | 0.56 |
ENSRNOT00000019874
|
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr10_+_12980674 | 0.55 |
ENSRNOT00000004638
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr14_-_18591394 | 0.52 |
ENSRNOT00000003716
|
Ereg
|
epiregulin |
| chr3_+_100769839 | 0.49 |
ENSRNOT00000077703
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_-_229601032 | 0.48 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr3_+_20965751 | 0.48 |
ENSRNOT00000010450
|
Olr417
|
olfactory receptor 417 |
| chr1_-_245514252 | 0.47 |
ENSRNOT00000033724
|
Pum3
|
pumilio RNA-binding family member 3 |
| chr1_+_98440186 | 0.45 |
ENSRNOT00000024150
|
Iglon5
|
IgLON family member 5 |
| chr1_+_213870502 | 0.42 |
ENSRNOT00000086483
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr20_+_4576057 | 0.41 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr8_+_57964988 | 0.40 |
ENSRNOT00000009497
|
Kdelc2
|
KDEL motif containing 2 |
| chr1_-_274106502 | 0.37 |
ENSRNOT00000020539
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr20_+_1861272 | 0.36 |
ENSRNOT00000044570
|
Olr1744
|
olfactory receptor 1744 |
| chr8_+_58347736 | 0.36 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr4_-_121822781 | 0.35 |
ENSRNOT00000072082
|
Vom1r95
|
vomeronasal 1 receptor 95 |
| chr19_+_37127508 | 0.33 |
ENSRNOT00000019656
|
Cbfb
|
core-binding factor, beta subunit |
| chr5_+_173256834 | 0.33 |
ENSRNOT00000089936
|
Ccnl2
|
cyclin L2 |
| chr14_+_19319299 | 0.30 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr7_-_105592804 | 0.30 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr3_-_176706896 | 0.28 |
ENSRNOT00000017337
|
Ptk6
|
protein tyrosine kinase 6 |
| chr5_-_145418992 | 0.25 |
ENSRNOT00000037128
|
Gjb4
|
gap junction protein, beta 4 |
| chr3_-_38741113 | 0.21 |
ENSRNOT00000006549
|
Prpf40a
|
pre-mRNA processing factor 40 homolog A |
| chr2_-_197935567 | 0.21 |
ENSRNOT00000085404
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr12_-_39882030 | 0.17 |
ENSRNOT00000078761
ENSRNOT00000048851 |
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr15_+_34520142 | 0.16 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chrX_-_156891038 | 0.15 |
ENSRNOT00000091495
|
Avpr2
|
arginine vasopressin receptor 2 |
| chr4_-_121811539 | 0.14 |
ENSRNOT00000049591
|
Vom1r94
|
vomeronasal 1 receptor 94 |
| chr1_-_229639187 | 0.13 |
ENSRNOT00000016804
|
Zfp91
|
zinc finger protein 91 |
| chr4_+_1671269 | 0.12 |
ENSRNOT00000071452
|
Olr1251
|
olfactory receptor 1251 |
| chr10_+_109107389 | 0.11 |
ENSRNOT00000068437
ENSRNOT00000005687 |
Baiap2
|
BAI1-associated protein 2 |
| chr8_+_40345160 | 0.07 |
ENSRNOT00000076308
|
Olr1200
|
olfactory receptor 1200 |
| chr7_+_13631975 | 0.06 |
ENSRNOT00000083548
|
Olr1084
|
olfactory receptor 1084 |
| chr3_-_103066392 | 0.04 |
ENSRNOT00000044181
|
Olr776
|
olfactory receptor 776 |
| chr14_-_17436897 | 0.04 |
ENSRNOT00000003277
|
Uso1
|
USO1 vesicle transport factor |
| chr7_-_135630654 | 0.02 |
ENSRNOT00000047388
ENSRNOT00000088223 ENSRNOT00000074793 |
Adamts20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr1_-_189911571 | 0.01 |
ENSRNOT00000080996
ENSRNOT00000088536 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr5_+_59348639 | 0.01 |
ENSRNOT00000084031
ENSRNOT00000060264 |
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chrX_-_33665821 | 0.01 |
ENSRNOT00000066676
|
Rbbp7
|
RB binding protein 7, chromatin remodeling factor |
| chr20_+_11863082 | 0.01 |
ENSRNOT00000001640
|
Fam207a
|
family with sequence similarity 207, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.5 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.2 | 6.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.8 | 7.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 1.4 | 4.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 1.2 | 4.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 1.2 | 4.9 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 1.1 | 3.4 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 1.0 | 3.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.9 | 5.7 | GO:1904714 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.7 | 4.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.6 | 4.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.6 | 1.8 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.6 | 1.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.5 | 1.6 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.5 | 1.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.5 | 3.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.4 | 1.6 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.4 | 3.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.3 | 1.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 3.4 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.3 | 5.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.3 | 3.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 2.4 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.3 | 4.1 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.2 | 1.0 | GO:0036166 | DNA methylation on cytosine within a CG sequence(GO:0010424) phenotypic switching(GO:0036166) |
| 0.2 | 1.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.2 | 1.7 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.2 | 5.3 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.2 | 0.6 | GO:0045575 | basophil activation(GO:0045575) |
| 0.2 | 1.1 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 1.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 4.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.2 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.1 | 0.8 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 4.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.5 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.6 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 2.0 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 1.9 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.9 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.5 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 10.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 12.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 6.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.6 | GO:0007631 | feeding behavior(GO:0007631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.0 | 7.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.0 | 4.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.8 | 3.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.7 | 6.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 4.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.9 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.6 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 0.8 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 2.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.6 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.6 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 1.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 4.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 4.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 12.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 5.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 4.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.8 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.8 | 2.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.7 | 7.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.6 | 5.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.6 | 4.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 1.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.4 | 3.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.4 | 1.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.4 | 1.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 1.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 4.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 3.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 3.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 2.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.7 | GO:0032407 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.1 | 11.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 4.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 4.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 4.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 8.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 12.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 6.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 3.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.6 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 1.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.8 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 3.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 8.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 4.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 4.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 3.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 4.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 5.1 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 3.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |