Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
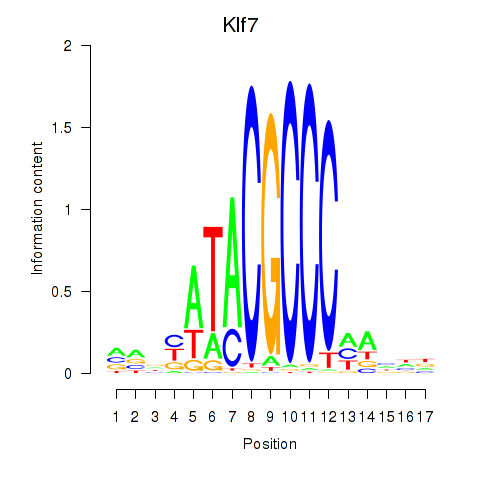
Results for Klf7
Z-value: 0.56
Transcription factors associated with Klf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf7
|
ENSRNOG00000046242 | Kruppel like factor 7 |
|
Klf7
|
ENSRNOG00000012961 | Kruppel like factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf7 | rn6_v1_chr9_-_70787913_70787913 | 0.07 | 2.3e-01 | Click! |
Activity profile of Klf7 motif
Sorted Z-values of Klf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_56962161 | 12.23 |
ENSRNOT00000026038
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr9_-_82419288 | 10.16 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr7_-_138039630 | 9.97 |
ENSRNOT00000008138
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr9_-_69953182 | 8.98 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr1_-_56683731 | 8.92 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr7_-_138039984 | 8.78 |
ENSRNOT00000089806
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr15_-_28313841 | 8.26 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr9_-_38196273 | 6.40 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr16_-_81028656 | 5.82 |
ENSRNOT00000091296
|
Dcun1d2
|
defective in cullin neddylation 1 domain containing 2 |
| chr4_-_120817377 | 5.70 |
ENSRNOT00000021826
|
Podxl2
|
podocalyxin-like 2 |
| chr10_+_58613674 | 5.48 |
ENSRNOT00000010975
|
Fam64a
|
family with sequence similarity 64, member A |
| chr20_+_3422461 | 4.98 |
ENSRNOT00000084917
ENSRNOT00000079854 |
Tubb5
|
tubulin, beta 5 class I |
| chr1_-_188190778 | 4.81 |
ENSRNOT00000092657
ENSRNOT00000022988 |
Coq7
|
coenzyme Q7, hydroxylase |
| chr2_+_113984824 | 4.23 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr7_-_134722215 | 4.22 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr16_-_74408030 | 4.22 |
ENSRNOT00000026418
|
Slc20a2
|
solute carrier family 20 member 2 |
| chr8_-_77992621 | 4.10 |
ENSRNOT00000085843
|
Myzap
|
myocardial zonula adherens protein |
| chr1_+_56242346 | 4.07 |
ENSRNOT00000019317
|
Smoc2
|
SPARC related modular calcium binding 2 |
| chr10_-_25890639 | 3.98 |
ENSRNOT00000085499
|
Hmmr
|
hyaluronan-mediated motility receptor |
| chr9_+_23420654 | 3.84 |
ENSRNOT00000073595
|
LOC688459
|
hypothetical protein LOC688459 |
| chr2_+_113984646 | 3.72 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr10_+_59173268 | 3.65 |
ENSRNOT00000013486
|
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr7_-_140640953 | 3.57 |
ENSRNOT00000083156
|
Tuba1a
|
tubulin, alpha 1A |
| chr1_+_213991620 | 3.56 |
ENSRNOT00000038691
|
Rplp2
|
ribosomal protein lateral stalk subunit P2 |
| chr16_+_36116258 | 3.53 |
ENSRNOT00000017652
|
Sap30
|
Sin3A associated protein 30 |
| chr13_-_87847263 | 3.49 |
ENSRNOT00000003650
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr1_+_220400855 | 3.38 |
ENSRNOT00000027178
|
Slc29a2
|
solute carrier family 29 member 2 |
| chr12_-_38274036 | 3.30 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr3_+_112428395 | 3.25 |
ENSRNOT00000079109
ENSRNOT00000048141 |
Stard9
|
StAR-related lipid transfer domain containing 9 |
| chr4_+_167219728 | 3.00 |
ENSRNOT00000075273
|
Smim10l1
|
small integral membrane protein 10 like 1 |
| chr1_+_220307394 | 2.87 |
ENSRNOT00000071891
|
LOC108348052
|
equilibrative nucleoside transporter 2 |
| chr8_+_72029489 | 2.82 |
ENSRNOT00000089336
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr7_+_140758615 | 2.69 |
ENSRNOT00000089448
|
Troap
|
trophinin associated protein |
| chr16_+_74408183 | 2.65 |
ENSRNOT00000036506
|
Smim19
|
small integral membrane protein 19 |
| chr10_+_86819929 | 2.56 |
ENSRNOT00000038228
|
Cdc6
|
cell division cycle 6 |
| chr11_+_44877859 | 2.54 |
ENSRNOT00000060838
|
Col8a1
|
collagen type VIII alpha 1 chain |
| chr7_-_145450233 | 2.54 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr16_-_47207487 | 2.53 |
ENSRNOT00000059459
|
Dctd
|
dCMP deaminase |
| chr10_-_13619935 | 2.45 |
ENSRNOT00000064392
|
Ccnf
|
cyclin F |
| chr19_+_52647070 | 2.43 |
ENSRNOT00000068389
ENSRNOT00000087857 |
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr3_-_79743737 | 2.42 |
ENSRNOT00000013584
|
Ptpmt1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr7_-_144322240 | 2.35 |
ENSRNOT00000089290
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr10_-_49192693 | 2.31 |
ENSRNOT00000004294
|
Zfp286a
|
zinc finger protein 286A |
| chr8_+_122003916 | 2.23 |
ENSRNOT00000091980
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr7_-_130176696 | 2.23 |
ENSRNOT00000051347
|
Dennd6b
|
DENN domain containing 6B |
| chr9_-_61033553 | 2.18 |
ENSRNOT00000002864
ENSRNOT00000082681 |
Gtf3c3
|
general transcription factor IIIC subunit 3 |
| chr7_+_144865608 | 2.18 |
ENSRNOT00000091596
ENSRNOT00000055285 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr12_-_39185168 | 2.03 |
ENSRNOT00000001799
|
Rnf34
|
ring finger protein 34 |
| chr1_-_60332899 | 2.03 |
ENSRNOT00000078636
|
LOC108348087
|
vomeronasal type-1 receptor 4-like |
| chr1_+_214428526 | 2.00 |
ENSRNOT00000057030
|
LOC100911575
|
60S acidic ribosomal protein P2-like |
| chr10_+_86819472 | 1.97 |
ENSRNOT00000081424
|
Cdc6
|
cell division cycle 6 |
| chr10_-_15211325 | 1.86 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr1_+_196095214 | 1.84 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chr1_-_228263198 | 1.83 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr9_+_69953440 | 1.82 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr10_+_57260680 | 1.81 |
ENSRNOT00000035445
|
Gp1ba
|
glycoprotein Ib platelet alpha subunit |
| chr12_+_22026075 | 1.81 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr9_+_64095978 | 1.73 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr2_-_189899325 | 1.71 |
ENSRNOT00000017561
|
Chtop
|
chromatin target of PRMT1 |
| chr11_-_67618704 | 1.70 |
ENSRNOT00000042208
|
Ccdc58
|
coiled-coil domain containing 58 |
| chr8_-_116856548 | 1.67 |
ENSRNOT00000043825
|
Rnf123
|
ring finger protein 123 |
| chr13_-_78521911 | 1.63 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr10_-_38985466 | 1.62 |
ENSRNOT00000010121
|
Il13
|
interleukin 13 |
| chr5_+_48561339 | 1.59 |
ENSRNOT00000010614
ENSRNOT00000091827 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr19_+_14392423 | 1.53 |
ENSRNOT00000018880
|
Tom1
|
target of myb1 membrane trafficking protein |
| chr3_-_104018861 | 1.53 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr8_+_128806129 | 1.52 |
ENSRNOT00000025225
|
Rpsa
|
ribosomal protein SA |
| chrX_+_120859968 | 1.47 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr9_-_65329066 | 1.44 |
ENSRNOT00000018284
|
Ppil3
|
peptidylprolyl isomerase like 3 |
| chr10_-_25847994 | 1.40 |
ENSRNOT00000082076
|
Mat2b
|
methionine adenosyltransferase 2B |
| chr10_+_103956019 | 1.38 |
ENSRNOT00000045875
|
Mrpl58
|
mitochondrial ribosomal protein L58 |
| chr9_+_90880614 | 1.37 |
ENSRNOT00000077859
ENSRNOT00000020705 ENSRNOT00000089142 |
Mff
|
mitochondrial fission factor |
| chr8_+_4019722 | 1.36 |
ENSRNOT00000073415
|
Vom2r23
|
vomeronasal 2 receptor, 23 |
| chrX_+_120860178 | 1.36 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr17_-_71897972 | 1.32 |
ENSRNOT00000065942
|
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr1_+_86948918 | 1.32 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr4_+_173115125 | 1.30 |
ENSRNOT00000074673
|
LOC100910575
|
serine-threonine kinase receptor-associated protein-like |
| chr3_-_165039707 | 1.21 |
ENSRNOT00000081648
|
Kcng1
|
potassium voltage-gated channel modifier subfamily G member 1 |
| chr1_+_178039063 | 1.20 |
ENSRNOT00000046313
|
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr9_-_64096265 | 1.15 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr8_+_42260375 | 1.12 |
ENSRNOT00000074713
|
LOC100912245
|
olfactory receptor 143-like |
| chr10_-_15161938 | 1.11 |
ENSRNOT00000026641
|
Fam173a
|
family with sequence similarity 173, member A |
| chr4_-_150616895 | 1.09 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr10_-_65805693 | 1.08 |
ENSRNOT00000012245
|
Tnfaip1
|
TNF alpha induced protein 1 |
| chr4_-_182844291 | 1.07 |
ENSRNOT00000064320
|
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr18_+_55505993 | 1.07 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr6_-_7741414 | 1.05 |
ENSRNOT00000038246
|
Thada
|
THADA, armadillo repeat containing |
| chr1_+_60117804 | 1.04 |
ENSRNOT00000080437
|
Vom1r8
|
vomeronasal 1 receptor 8 |
| chr1_-_52992213 | 1.04 |
ENSRNOT00000033528
|
Prr18
|
proline rich 18 |
| chr8_-_114449956 | 1.01 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr17_-_18028808 | 0.98 |
ENSRNOT00000022244
|
Kdm1b
|
lysine demethylase 1B |
| chr5_+_107233230 | 0.95 |
ENSRNOT00000029976
|
Ifne
|
interferon, epsilon |
| chr19_-_29802083 | 0.94 |
ENSRNOT00000024755
|
AC123471.1
|
|
| chr12_+_38274297 | 0.94 |
ENSRNOT00000087905
ENSRNOT00000057788 |
Rsrc2
|
arginine and serine rich coiled-coil 2 |
| chr1_+_168378637 | 0.91 |
ENSRNOT00000028936
|
Olr86
|
olfactory receptor 86 |
| chr2_+_150211898 | 0.90 |
ENSRNOT00000018767
|
Sucnr1
|
succinate receptor 1 |
| chr14_+_100415668 | 0.89 |
ENSRNOT00000008057
|
C1d
|
C1D nuclear receptor co-repressor |
| chr1_-_100810522 | 0.89 |
ENSRNOT00000027260
|
Atf5
|
activating transcription factor 5 |
| chr2_-_35435719 | 0.88 |
ENSRNOT00000087827
|
LOC100910041
|
olfactory receptor 146-like |
| chr14_-_34218961 | 0.86 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr15_-_36213666 | 0.85 |
ENSRNOT00000078704
|
Olr1291
|
olfactory receptor 1291 |
| chr8_-_43007390 | 0.83 |
ENSRNOT00000072355
|
LOC100910445
|
olfactory receptor 146-like |
| chr15_+_28377996 | 0.77 |
ENSRNOT00000078475
|
Arhgef40
|
Rho guanine nucleotide exchange factor 40 |
| chr8_+_49544858 | 0.75 |
ENSRNOT00000075133
|
LOC102546809
|
uncharacterized LOC102546809 |
| chr9_-_23352668 | 0.75 |
ENSRNOT00000075279
|
Mut
|
methylmalonyl CoA mutase |
| chr4_+_7835949 | 0.75 |
ENSRNOT00000065962
|
Tomm7
|
translocase of outer mitochondrial membrane 7 |
| chr18_+_55391388 | 0.75 |
ENSRNOT00000071612
|
LOC100910934
|
interferon-inducible GTPase 1-like |
| chr11_-_81379640 | 0.68 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr18_+_59748444 | 0.66 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr17_-_62462644 | 0.63 |
ENSRNOT00000024545
|
Ccny
|
cyclin Y |
| chr19_-_28296640 | 0.62 |
ENSRNOT00000041115
ENSRNOT00000042781 |
AABR07043429.1
|
|
| chr1_+_85470831 | 0.61 |
ENSRNOT00000057096
|
Timm50
|
translocase of inner mitochondrial membrane 50 |
| chr4_-_28310178 | 0.58 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr11_-_81379871 | 0.57 |
ENSRNOT00000089294
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr1_-_226630900 | 0.49 |
ENSRNOT00000028092
|
Tmem138
|
transmembrane protein 138 |
| chr2_-_35485401 | 0.49 |
ENSRNOT00000072751
|
AABR07007931.1
|
|
| chr9_-_69878706 | 0.46 |
ENSRNOT00000035879
|
Ino80d
|
INO80 complex subunit D |
| chr10_+_44804631 | 0.45 |
ENSRNOT00000040720
|
Olr1460
|
olfactory receptor 1460 |
| chr10_-_89225297 | 0.45 |
ENSRNOT00000027868
ENSRNOT00000082010 |
Becn1
|
beclin 1 |
| chr1_-_22508170 | 0.33 |
ENSRNOT00000050763
|
Taar7c
|
trace amine-associated receptor 7C |
| chr12_-_17972737 | 0.21 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr2_+_12102487 | 0.18 |
ENSRNOT00000089209
ENSRNOT00000072155 |
LOC100912538
|
centrin-3-like |
| chr3_+_150055749 | 0.12 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr5_+_126254142 | 0.10 |
ENSRNOT00000057471
|
Pars2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 18.8 | GO:0006868 | glutamine transport(GO:0006868) |
| 2.4 | 12.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 2.1 | 8.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.6 | 4.8 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 1.1 | 3.4 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.7 | 3.6 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.6 | 4.5 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.6 | 1.7 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.6 | 8.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.5 | 2.2 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.5 | 1.6 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.5 | 1.6 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.5 | 2.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.5 | 1.5 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.5 | 4.2 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.4 | 1.3 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.4 | 2.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.4 | 6.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 2.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.3 | 1.4 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.3 | 1.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 2.9 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.3 | 5.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 1.2 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.2 | 2.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.7 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 5.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 1.7 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.2 | 0.5 | GO:0032258 | CVT pathway(GO:0032258) |
| 0.1 | 9.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 2.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.7 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 4.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 8.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 2.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.1 | 1.9 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 3.6 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.0 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 8.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 1.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.9 | GO:0021930 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.1 | 2.5 | GO:0006220 | pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 2.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 3.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 4.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 1.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.9 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.5 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 1.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 2.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.6 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 1.6 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 7.5 | GO:0007017 | microtubule-based process(GO:0007017) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.1 | 3.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.7 | 2.2 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.7 | 3.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.6 | 1.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 4.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 5.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 8.9 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.3 | 2.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 2.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 4.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 9.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 0.5 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 10.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 2.6 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 3.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 4.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 8.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 13.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 15.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 6.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 3.1 | 12.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.8 | 4.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.5 | 2.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 1.6 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.4 | 1.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.4 | 8.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 9.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 1.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 5.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 2.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 1.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 2.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.3 | 2.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 5.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 3.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 1.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 4.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 13.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 3.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.5 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.7 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 10.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 8.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 3.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 7.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 6.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.6 | GO:0005125 | cytokine activity(GO:0005125) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 8.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 12.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 8.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 4.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 8.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 6.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 12.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 4.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 4.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 1.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 9.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 2.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 8.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 4.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 5.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 5.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |