Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
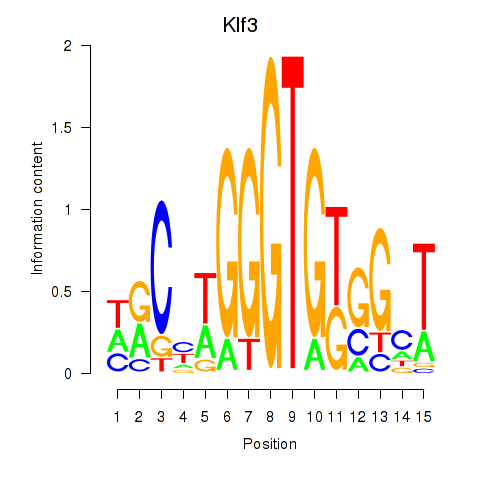
Results for Klf3
Z-value: 0.19
Transcription factors associated with Klf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf3
|
ENSRNOG00000002163 | Kruppel like factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf3 | rn6_v1_chr14_-_45165207_45165207 | 0.21 | 1.8e-04 | Click! |
Activity profile of Klf3 motif
Sorted Z-values of Klf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_7064870 | 4.03 |
ENSRNOT00000019983
|
Stx11
|
syntaxin 11 |
| chr1_+_226091774 | 2.67 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr4_+_100407658 | 1.89 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chr8_-_94563760 | 1.69 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
| chr19_-_1074333 | 1.68 |
ENSRNOT00000017983
ENSRNOT00000086995 |
Cdh5
|
cadherin 5 |
| chrX_-_72133692 | 1.32 |
ENSRNOT00000004263
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr10_-_56465393 | 1.16 |
ENSRNOT00000056860
|
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr8_-_117082162 | 1.03 |
ENSRNOT00000075241
|
Tcta
|
T-cell leukemia translocation altered |
| chr2_+_206314213 | 0.94 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr7_-_144322240 | 0.79 |
ENSRNOT00000089290
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr7_-_145450233 | 0.74 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr7_-_75597087 | 0.61 |
ENSRNOT00000080676
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr13_+_27449934 | 0.51 |
ENSRNOT00000003409
|
Serpinb2
|
serpin family B member 2 |
| chr1_-_78968329 | 0.47 |
ENSRNOT00000023078
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr8_-_55171718 | 0.46 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr8_+_118013612 | 0.20 |
ENSRNOT00000056166
|
Map4
|
microtubule-associated protein 4 |
| chr1_-_274106502 | 0.17 |
ENSRNOT00000020539
|
Smndc1
|
survival motor neuron domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) |
| 0.3 | 1.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 1.3 | GO:0071104 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) |
| 0.2 | 1.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 2.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 1.2 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0098830 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.3 | 1.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 4.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 4.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |