Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
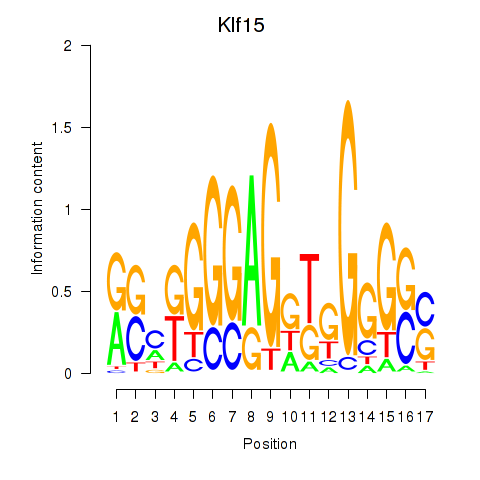
Results for Klf15
Z-value: 0.74
Transcription factors associated with Klf15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf15
|
ENSRNOG00000017808 | Kruppel-like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf15 | rn6_v1_chr4_+_122365093_122365093 | 0.35 | 1.1e-10 | Click! |
Activity profile of Klf15 motif
Sorted Z-values of Klf15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_3657325 | 18.33 |
ENSRNOT00000010927
|
Tmem178a
|
transmembrane protein 178A |
| chr13_-_74077783 | 17.64 |
ENSRNOT00000005677
|
Soat1
|
sterol O-acyltransferase 1 |
| chr20_+_4363508 | 16.58 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chrX_+_112769645 | 16.51 |
ENSRNOT00000064478
|
Col4a5
|
collagen type IV alpha 5 chain |
| chr20_+_4363152 | 15.73 |
ENSRNOT00000000508
ENSRNOT00000084841 ENSRNOT00000072848 ENSRNOT00000077561 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr9_+_88357556 | 15.66 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
| chr10_+_90550147 | 14.85 |
ENSRNOT00000032944
|
Fzd2
|
frizzled class receptor 2 |
| chr9_-_88357182 | 14.83 |
ENSRNOT00000041176
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr12_-_22417980 | 14.05 |
ENSRNOT00000072208
|
Ephb4
|
EPH receptor B4 |
| chr5_-_147412705 | 14.03 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr9_+_80118029 | 13.14 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr10_+_84152152 | 12.15 |
ENSRNOT00000010724
|
Hoxb5
|
homeo box B5 |
| chr2_-_206222248 | 12.01 |
ENSRNOT00000026075
|
Olfml3
|
olfactomedin-like 3 |
| chr1_-_263803150 | 11.32 |
ENSRNOT00000017840
|
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr20_+_48335540 | 11.14 |
ENSRNOT00000000352
|
Cd24
|
CD24 molecule |
| chr16_-_6404578 | 9.99 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr14_-_82287706 | 9.77 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr2_+_225005019 | 9.50 |
ENSRNOT00000015579
|
Cnn3
|
calponin 3 |
| chr18_-_47513030 | 9.42 |
ENSRNOT00000083881
ENSRNOT00000074226 |
Lox
|
lysyl oxidase |
| chr20_+_41266566 | 9.41 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr6_-_99870024 | 9.19 |
ENSRNOT00000010043
|
Rab15
|
RAB15, member RAS oncogene family |
| chr6_+_33885495 | 9.12 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr15_+_34520142 | 8.48 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr1_-_170628915 | 7.86 |
ENSRNOT00000042865
|
Dchs1
|
dachsous cadherin-related 1 |
| chr16_-_20592449 | 7.78 |
ENSRNOT00000026821
|
Isyna1
|
inositol-3-phosphate synthase 1 |
| chr1_+_101610773 | 7.20 |
ENSRNOT00000032535
|
Rasip1
|
Ras interacting protein 1 |
| chr15_+_34493138 | 7.00 |
ENSRNOT00000089584
ENSRNOT00000027789 |
Nfatc4
|
nuclear factor of activated T-cells 4 |
| chr1_+_226435979 | 6.91 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr1_+_78800754 | 6.91 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr15_+_120372 | 6.85 |
ENSRNOT00000007828
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr5_+_166533181 | 6.79 |
ENSRNOT00000045063
|
Clstn1
|
calsyntenin 1 |
| chr1_+_80000165 | 6.07 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr6_-_122721496 | 6.04 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr9_+_93326283 | 6.03 |
ENSRNOT00000024578
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr3_+_110734105 | 5.71 |
ENSRNOT00000073587
|
Chst14
|
carbohydrate sulfotransferase 14 |
| chr1_+_83653234 | 5.24 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr6_+_126018841 | 5.11 |
ENSRNOT00000089840
ENSRNOT00000088089 |
Slc24a4
|
solute carrier family 24 member 4 |
| chr20_-_5064469 | 4.93 |
ENSRNOT00000001120
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr1_+_221099998 | 4.82 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr19_+_49792273 | 4.82 |
ENSRNOT00000017941
ENSRNOT00000083991 |
Cmip
|
c-Maf-inducing protein |
| chr5_+_43603043 | 4.78 |
ENSRNOT00000009899
|
Epha7
|
Eph receptor A7 |
| chr13_-_83842051 | 4.67 |
ENSRNOT00000004376
|
Mpzl1
|
myelin protein zero-like 1 |
| chr20_+_5646097 | 4.57 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr9_+_93545396 | 4.53 |
ENSRNOT00000025093
|
LOC100359583
|
hypothetical protein LOC100359583 |
| chr6_-_103470427 | 4.53 |
ENSRNOT00000091560
ENSRNOT00000088795 ENSRNOT00000079824 |
Actn1
|
actinin, alpha 1 |
| chr6_-_3444519 | 4.51 |
ENSRNOT00000010366
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr1_+_282265370 | 4.42 |
ENSRNOT00000015687
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr7_-_90318221 | 4.42 |
ENSRNOT00000050774
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr13_+_78979321 | 4.27 |
ENSRNOT00000003857
|
Ankrd45
|
ankyrin repeat domain 45 |
| chr7_+_97559841 | 4.21 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr11_-_38457373 | 4.18 |
ENSRNOT00000041177
|
Zfp295
|
zinc finger protein 295 |
| chr4_-_66002444 | 4.16 |
ENSRNOT00000018645
|
Zc3hav1l
|
zinc finger CCCH-type containing, antiviral 1 like |
| chrX_-_22440187 | 4.15 |
ENSRNOT00000090731
|
Gpr173
|
|
| chr9_-_16979044 | 4.13 |
ENSRNOT00000073191
ENSRNOT00000025335 |
Zfp318
|
zinc finger protein 318 |
| chr5_-_172623899 | 3.94 |
ENSRNOT00000080591
|
Ski
|
SKI proto-oncogene |
| chr8_+_117068582 | 3.83 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr16_+_20740826 | 3.69 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chrX_+_70596901 | 3.61 |
ENSRNOT00000088114
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr3_+_58632476 | 3.56 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chr8_+_117780891 | 3.52 |
ENSRNOT00000077236
|
Shisa5
|
shisa family member 5 |
| chr5_-_100647727 | 3.50 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr14_+_83510278 | 3.42 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr2_+_207923775 | 3.40 |
ENSRNOT00000019997
ENSRNOT00000051835 |
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr2_+_198823366 | 3.38 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr5_-_100647298 | 3.35 |
ENSRNOT00000067538
ENSRNOT00000013092 |
Nfib
|
nuclear factor I/B |
| chr18_+_72005581 | 3.32 |
ENSRNOT00000072519
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr1_+_226091774 | 3.30 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr1_-_199395363 | 3.22 |
ENSRNOT00000090368
ENSRNOT00000026587 |
Prss36
|
protease, serine, 36 |
| chr1_+_225096598 | 3.20 |
ENSRNOT00000035650
|
Ganab
|
glucosidase II alpha subunit |
| chr5_+_74649765 | 3.11 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chr7_+_123381077 | 3.10 |
ENSRNOT00000082603
ENSRNOT00000056041 |
Srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr13_-_52256196 | 3.07 |
ENSRNOT00000011105
|
Ipo9
|
importin 9 |
| chr8_-_55408464 | 2.95 |
ENSRNOT00000066893
|
Sik2
|
salt-inducible kinase 2 |
| chr20_+_5526975 | 2.92 |
ENSRNOT00000059548
|
Phf1
|
PHD finger protein 1 |
| chr8_+_117737117 | 2.91 |
ENSRNOT00000028039
|
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr4_+_10631073 | 2.89 |
ENSRNOT00000081939
ENSRNOT00000017907 |
Ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr7_-_139318455 | 2.83 |
ENSRNOT00000092029
|
Hdac7
|
histone deacetylase 7 |
| chr5_+_78483893 | 2.80 |
ENSRNOT00000059183
ENSRNOT00000059181 |
Rgs3
|
regulator of G-protein signaling 3 |
| chrX_-_118615798 | 2.74 |
ENSRNOT00000045463
|
Lrch2
|
leucine rich repeats and calponin homology domain containing 2 |
| chrX_-_157172068 | 2.71 |
ENSRNOT00000087962
|
Dusp9
|
dual specificity phosphatase 9 |
| chr6_-_43493816 | 2.62 |
ENSRNOT00000077786
|
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr9_+_61720583 | 2.60 |
ENSRNOT00000020536
|
Mob4
|
MOB family member 4, phocein |
| chr6_+_105364668 | 2.50 |
ENSRNOT00000009513
ENSRNOT00000087090 |
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr20_+_5527181 | 2.40 |
ENSRNOT00000091364
|
Phf1
|
PHD finger protein 1 |
| chr19_+_25181564 | 2.34 |
ENSRNOT00000008104
|
Rfx1
|
regulatory factor X1 |
| chr8_+_117737387 | 2.29 |
ENSRNOT00000090164
ENSRNOT00000091573 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr1_+_220992770 | 2.25 |
ENSRNOT00000045233
|
Rela
|
RELA proto-oncogene, NF-kB subunit |
| chr10_-_46332172 | 2.21 |
ENSRNOT00000004475
|
Rasd1
|
ras related dexamethasone induced 1 |
| chr7_+_130474279 | 2.18 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr17_-_85141210 | 2.16 |
ENSRNOT00000000162
|
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr10_+_59585072 | 2.14 |
ENSRNOT00000025009
|
Camkk1
|
calcium/calmodulin-dependent protein kinase kinase 1 |
| chr19_-_645937 | 2.06 |
ENSRNOT00000016521
|
Car7
|
carbonic anhydrase 7 |
| chr8_+_45797315 | 2.05 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr14_+_60857989 | 2.01 |
ENSRNOT00000034411
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr10_+_66690133 | 1.96 |
ENSRNOT00000046262
|
Nf1
|
neurofibromin 1 |
| chr6_-_75670135 | 1.93 |
ENSRNOT00000007300
|
Snx6
|
sorting nexin 6 |
| chr7_-_77162148 | 1.91 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr2_+_189615948 | 1.87 |
ENSRNOT00000092144
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr1_-_190965115 | 1.83 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr7_+_130474508 | 1.79 |
ENSRNOT00000085191
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr3_-_164095878 | 1.71 |
ENSRNOT00000079414
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chr4_-_115157263 | 1.68 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr6_-_26685235 | 1.67 |
ENSRNOT00000066743
|
Atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr3_-_91839009 | 1.66 |
ENSRNOT00000083703
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr6_-_26771164 | 1.64 |
ENSRNOT00000009090
|
Tcf23
|
transcription factor 23 |
| chr1_+_190964885 | 1.50 |
ENSRNOT00000039186
|
Mettl9
|
methyltransferase like 9 |
| chr11_+_24263281 | 1.43 |
ENSRNOT00000086946
|
Gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr10_+_59360765 | 1.38 |
ENSRNOT00000036278
|
Zzef1
|
zinc finger ZZ-type and EF-hand domain containing 1 |
| chr20_+_4576514 | 1.31 |
ENSRNOT00000090125
ENSRNOT00000047370 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr19_-_41798383 | 1.24 |
ENSRNOT00000021744
|
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr9_+_99720856 | 1.21 |
ENSRNOT00000047487
|
Olr1346
|
olfactory receptor 1346 |
| chr10_-_19164505 | 1.20 |
ENSRNOT00000009058
|
Foxi1
|
forkhead box I1 |
| chr13_+_49660356 | 1.01 |
ENSRNOT00000012844
|
Lrrn2
|
leucine rich repeat neuronal 2 |
| chr5_-_82168347 | 0.99 |
ENSRNOT00000084959
ENSRNOT00000084147 |
Astn2
|
astrotactin 2 |
| chr8_+_20230082 | 0.95 |
ENSRNOT00000044463
|
Olr1165
|
olfactory receptor 1165 |
| chr16_-_36373546 | 0.93 |
ENSRNOT00000079552
|
Hand2
|
heart and neural crest derivatives expressed transcript 2 |
| chr6_-_115352681 | 0.85 |
ENSRNOT00000005873
|
Gtf2a1
|
general transcription factor 2A subunit 1 |
| chr8_+_128087345 | 0.84 |
ENSRNOT00000019777
|
Acvr2b
|
activin A receptor type 2B |
| chr10_-_13542077 | 0.82 |
ENSRNOT00000008736
|
Atp6v0c
|
ATPase H+ transporting V0 subunit C |
| chr2_-_94730978 | 0.82 |
ENSRNOT00000087598
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr18_-_5314511 | 0.79 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr7_+_122818975 | 0.78 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chr16_+_60925093 | 0.78 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr7_+_144014173 | 0.78 |
ENSRNOT00000019403
|
Sp1
|
Sp1 transcription factor |
| chr3_-_7422738 | 0.75 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr20_+_4576057 | 0.69 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr11_+_71533078 | 0.58 |
ENSRNOT00000002405
|
Slc51a
|
solute carrier family 51, alpha subunit |
| chr10_-_64862268 | 0.57 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chrX_-_107542510 | 0.56 |
ENSRNOT00000074140
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr10_-_59360661 | 0.51 |
ENSRNOT00000036942
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr10_+_103972888 | 0.49 |
ENSRNOT00000067838
|
Kctd2
|
potassium channel tetramerization domain containing 2 |
| chr1_-_80367313 | 0.49 |
ENSRNOT00000023392
ENSRNOT00000084882 |
Mark4
|
microtubule affinity regulating kinase 4 |
| chr8_+_65686648 | 0.44 |
ENSRNOT00000017233
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr5_-_169167831 | 0.35 |
ENSRNOT00000012407
|
Phf13
|
PHD finger protein 13 |
| chr20_-_32052855 | 0.28 |
ENSRNOT00000074989
|
LOC100364027
|
hexokinase 1-like |
| chr3_-_2727616 | 0.07 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr7_-_14302552 | 0.06 |
ENSRNOT00000091368
|
Brd4
|
bromodomain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 32.3 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 5.0 | 14.9 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 3.9 | 7.9 | GO:0003192 | mitral valve formation(GO:0003192) |
| 3.7 | 11.1 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 3.5 | 14.1 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 2.9 | 17.6 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 2.3 | 30.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 2.3 | 7.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.3 | 6.9 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 2.3 | 9.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 2.0 | 9.8 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.8 | 3.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 1.7 | 6.9 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.7 | 6.8 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 1.6 | 4.8 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.4 | 8.5 | GO:0015074 | DNA integration(GO:0015074) |
| 1.1 | 16.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.0 | 7.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 1.0 | 3.8 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.9 | 9.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.9 | 3.7 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.9 | 10.0 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.8 | 4.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.8 | 4.0 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.8 | 3.9 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.8 | 3.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.8 | 4.6 | GO:0007223 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Wnt signaling pathway, calcium modulating pathway(GO:0007223) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) sensory perception of umami taste(GO:0050917) |
| 0.8 | 5.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.8 | 2.3 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.7 | 5.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.7 | 20.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.7 | 5.7 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.6 | 9.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.6 | 4.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.5 | 4.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.5 | 6.1 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.5 | 2.0 | GO:0036166 | DNA methylation on cytosine within a CG sequence(GO:0010424) phenotypic switching(GO:0036166) |
| 0.5 | 6.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 11.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.5 | 13.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.5 | 3.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.4 | 1.7 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.4 | 2.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 3.4 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.4 | 5.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 4.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 3.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 4.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 1.9 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.3 | 2.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 0.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 0.8 | GO:0018076 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 0.8 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 0.8 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.2 | 4.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 3.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.2 | 1.0 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 5.1 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 9.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 4.9 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 1.7 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 1.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.4 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 7.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 6.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 2.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.8 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.8 | GO:0048617 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 6.6 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.1 | 2.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 2.9 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 6.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 7.2 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 2.9 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 2.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 2.6 | GO:0006900 | membrane budding(GO:0006900) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 3.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 3.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.7 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 3.9 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.2 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.2 | GO:0021766 | hippocampus development(GO:0021766) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 47.0 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 1.0 | 3.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 4.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.8 | 10.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.7 | 3.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.7 | 6.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.6 | 6.9 | GO:0032009 | early phagosome(GO:0032009) |
| 0.6 | 11.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.6 | 32.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 1.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.3 | 5.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 2.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 4.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 3.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 2.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 9.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 3.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 12.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 5.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 4.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 4.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 60.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 15.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 2.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 16.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 6.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 9.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 15.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.6 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.0 | 6.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 32.3 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.9 | 9.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.5 | 13.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.4 | 5.6 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 1.3 | 7.5 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.2 | 9.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.0 | 5.7 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.9 | 18.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.9 | 14.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.8 | 4.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 4.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.7 | 5.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.7 | 3.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.6 | 17.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.6 | 40.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.6 | 4.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 5.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 16.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.5 | 11.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 3.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.5 | 6.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.5 | 5.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.5 | 9.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.5 | 2.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.4 | 1.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 3.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 4.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 7.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 4.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 12.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 2.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 2.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 8.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 4.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 3.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 7.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 2.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 3.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 4.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 2.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 6.0 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.2 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 9.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 6.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 7.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 3.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 4.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 4.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 10.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 21.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 1.1 | 56.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.6 | 30.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.5 | 14.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.5 | 16.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 10.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 4.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 4.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 7.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 4.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 8.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 2.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 3.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 6.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 7.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 18.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 9.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 32.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.0 | 44.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.8 | 9.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.6 | 13.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 9.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 4.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 6.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.3 | 4.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 3.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 2.1 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.2 | 4.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 7.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 5.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 5.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 13.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 11.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 3.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 5.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 3.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |