Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
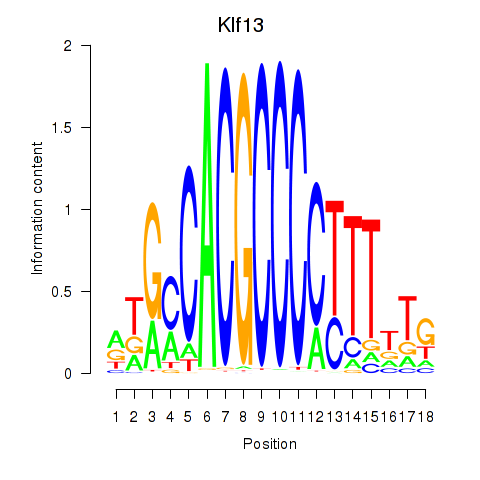
Results for Klf13
Z-value: 0.74
Transcription factors associated with Klf13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf13
|
ENSRNOG00000015822 | Kruppel-like factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf13 | rn6_v1_chr1_-_124803363_124803363 | -0.04 | 4.5e-01 | Click! |
Activity profile of Klf13 motif
Sorted Z-values of Klf13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_5049496 | 18.46 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr20_-_5155293 | 18.38 |
ENSRNOT00000092322
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr6_-_39363367 | 17.60 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr5_+_64476317 | 12.91 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr5_+_29538380 | 12.49 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr9_+_113699170 | 11.73 |
ENSRNOT00000017915
|
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr19_+_26106838 | 11.41 |
ENSRNOT00000035987
|
Hook2
|
hook microtubule-tethering protein 2 |
| chr4_-_117153907 | 11.09 |
ENSRNOT00000091374
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr19_+_10563423 | 9.95 |
ENSRNOT00000021037
|
Dok4
|
docking protein 4 |
| chr5_+_138470069 | 9.93 |
ENSRNOT00000076343
ENSRNOT00000064073 |
Zmynd12
|
zinc finger, MYND-type containing 12 |
| chr16_-_19894591 | 9.37 |
ENSRNOT00000085940
|
Ano8
|
anoctamin 8 |
| chr5_-_159583049 | 9.28 |
ENSRNOT00000055839
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr12_-_44279002 | 9.17 |
ENSRNOT00000064900
|
Fbxo21
|
F-box protein 21 |
| chr5_+_159428515 | 8.49 |
ENSRNOT00000010183
|
Padi2
|
peptidyl arginine deiminase 2 |
| chr12_+_22026075 | 8.37 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr1_-_101500850 | 8.31 |
ENSRNOT00000028390
|
Nucb1
|
nucleobindin 1 |
| chr5_+_154489590 | 8.23 |
ENSRNOT00000035788
|
Id3
|
inhibitor of DNA binding 3, HLH protein |
| chr12_-_24046814 | 8.20 |
ENSRNOT00000001961
|
Por
|
cytochrome p450 oxidoreductase |
| chr7_+_12619774 | 7.71 |
ENSRNOT00000015257
|
Med16
|
mediator complex subunit 16 |
| chr3_+_2480232 | 7.47 |
ENSRNOT00000014489
|
Tprn
|
taperin |
| chr9_+_66335492 | 7.22 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr1_+_80092403 | 7.10 |
ENSRNOT00000078336
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr1_+_80141630 | 6.82 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr13_+_41883137 | 6.45 |
ENSRNOT00000004581
|
Slc35f5
|
solute carrier family 35, member F5 |
| chr1_+_166739532 | 5.90 |
ENSRNOT00000079846
ENSRNOT00000026665 |
Clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr4_+_115473811 | 5.81 |
ENSRNOT00000042875
|
Nagk
|
N-acetylglucosamine kinase |
| chr10_-_64550145 | 5.63 |
ENSRNOT00000050232
|
Nxn
|
nucleoredoxin |
| chr1_+_266530477 | 5.58 |
ENSRNOT00000054699
|
Cnnm2
|
cyclin and CBS domain divalent metal cation transport mediator 2 |
| chr19_+_57047830 | 5.25 |
ENSRNOT00000080860
|
Galnt2
|
polypeptide N-acetylgalactosaminyltransferase 2 |
| chr8_+_12823155 | 5.17 |
ENSRNOT00000011008
|
Sesn3
|
sestrin 3 |
| chr5_-_58484900 | 5.11 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr4_-_116786391 | 4.81 |
ENSRNOT00000086297
ENSRNOT00000091490 |
Exoc6b
|
exocyst complex component 6B |
| chr20_-_10912769 | 4.68 |
ENSRNOT00000051678
|
RGD1566085
|
similar to pyridoxal (pyridoxine, vitamin B6) kinase |
| chr2_+_122877286 | 4.67 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr2_-_232117134 | 4.65 |
ENSRNOT00000030798
|
Alpk1
|
alpha-kinase 1 |
| chr1_+_89017479 | 4.54 |
ENSRNOT00000038154
|
U2af1l4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr2_+_28460068 | 4.50 |
ENSRNOT00000066819
|
Foxd1
|
forkhead box D1 |
| chr3_-_8432593 | 4.37 |
ENSRNOT00000090574
|
AC114363.1
|
|
| chr4_+_7661558 | 4.26 |
ENSRNOT00000080229
|
Fam126a
|
family with sequence similarity 126, member A |
| chr10_+_90230441 | 4.12 |
ENSRNOT00000082722
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr12_+_2069959 | 3.99 |
ENSRNOT00000001298
|
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr10_-_46145548 | 3.88 |
ENSRNOT00000033483
|
Pld6
|
phospholipase D family, member 6 |
| chr5_-_150506871 | 3.86 |
ENSRNOT00000086131
|
Trnau1ap
|
tRNA selenocysteine 1 associated protein 1 |
| chr11_+_42858478 | 3.65 |
ENSRNOT00000002293
|
Arl6
|
ADP-ribosylation factor like GTPase 6 |
| chr8_+_64364741 | 3.61 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr13_-_79801561 | 3.60 |
ENSRNOT00000075936
|
Suco
|
SUN domain containing ossification factor |
| chr12_-_39641285 | 3.57 |
ENSRNOT00000001729
|
Anapc7
|
anaphase promoting complex subunit 7 |
| chr20_-_3819200 | 3.40 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr12_-_19599374 | 3.32 |
ENSRNOT00000001849
|
Gpc2
|
glypican 2 |
| chr17_+_15845931 | 3.22 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr15_+_33069382 | 3.02 |
ENSRNOT00000060193
|
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr4_+_7662019 | 2.79 |
ENSRNOT00000014023
|
Fam126a
|
family with sequence similarity 126, member A |
| chr1_-_89017290 | 2.78 |
ENSRNOT00000028438
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr9_-_81879211 | 2.74 |
ENSRNOT00000085140
|
Rnf25
|
ring finger protein 25 |
| chr12_+_2070365 | 2.70 |
ENSRNOT00000072952
|
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr3_-_130114770 | 2.56 |
ENSRNOT00000010638
|
Jag1
|
jagged 1 |
| chr7_+_31784438 | 2.54 |
ENSRNOT00000010914
ENSRNOT00000010929 |
Ikbip
|
IKBKB interacting protein |
| chr1_-_19376301 | 2.45 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr13_-_79801368 | 2.44 |
ENSRNOT00000075998
ENSRNOT00000084058 |
Suco
|
SUN domain containing ossification factor |
| chr13_-_79801112 | 2.43 |
ENSRNOT00000087323
ENSRNOT00000036483 |
Suco
|
SUN domain containing ossification factor |
| chr1_-_78212350 | 2.40 |
ENSRNOT00000071098
|
Inafm1
|
InaF-motif containing 1 |
| chr12_+_19599834 | 2.31 |
ENSRNOT00000092039
ENSRNOT00000042006 |
Stag3
|
stromal antigen 3 |
| chr4_-_51726212 | 2.18 |
ENSRNOT00000009261
|
Wasl
|
Wiskott-Aldrich syndrome-like |
| chr3_-_60795951 | 2.15 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr2_-_260148589 | 2.15 |
ENSRNOT00000013238
|
Acadm
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr4_-_121565212 | 2.13 |
ENSRNOT00000088753
|
Chchd6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr6_-_102353403 | 2.11 |
ENSRNOT00000090407
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr13_+_44957014 | 2.01 |
ENSRNOT00000004878
|
Ubxn4
|
UBX domain protein 4 |
| chr20_-_5533600 | 1.85 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr7_+_11414446 | 1.77 |
ENSRNOT00000027441
|
Pias4
|
protein inhibitor of activated STAT, 4 |
| chr20_-_5533448 | 1.77 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr13_-_102643223 | 1.76 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr11_+_87522971 | 1.58 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr13_-_113817995 | 1.44 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chr1_-_261371508 | 1.43 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr10_+_56576428 | 1.42 |
ENSRNOT00000079237
ENSRNOT00000023291 |
Cldn7
|
claudin 7 |
| chr9_-_43022998 | 1.37 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr1_+_198210525 | 1.35 |
ENSRNOT00000026755
|
Ypel3
|
yippee-like 3 |
| chr1_-_164814651 | 1.31 |
ENSRNOT00000024420
|
Neu3
|
neuraminidase 3 |
| chr1_-_78573374 | 1.30 |
ENSRNOT00000090519
|
Arhgap35
|
Rho GTPase activating protein 35 |
| chr6_-_108329464 | 1.19 |
ENSRNOT00000016040
|
Abcd4
|
ATP binding cassette subfamily D member 4 |
| chr10_-_11035484 | 0.91 |
ENSRNOT00000077164
|
Hmox2
|
heme oxygenase 2 |
| chr10_+_93354003 | 0.89 |
ENSRNOT00000008140
|
Mettl2b
|
methyltransferase like 2B |
| chr4_-_113497396 | 0.80 |
ENSRNOT00000079173
ENSRNOT00000067314 |
Pole4
|
DNA polymerase epsilon 4, accessory subunit |
| chr20_+_3351303 | 0.72 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr20_+_10265806 | 0.65 |
ENSRNOT00000001564
ENSRNOT00000086272 |
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chrX_-_65335987 | 0.52 |
ENSRNOT00000047128
|
AABR07038981.1
|
|
| chr3_+_164274710 | 0.38 |
ENSRNOT00000012939
|
Snai1
|
snail family transcriptional repressor 1 |
| chr7_+_117605050 | 0.23 |
ENSRNOT00000047380
|
Slc52a2
|
solute carrier family 52 member 2 |
| chr20_-_49486550 | 0.07 |
ENSRNOT00000048270
ENSRNOT00000076541 |
Prdm1
|
PR/SET domain 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 3.1 | 9.3 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 2.1 | 8.5 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 2.1 | 8.2 | GO:0042126 | growth plate cartilage chondrocyte proliferation(GO:0003419) nitrate metabolic process(GO:0042126) |
| 2.0 | 11.7 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.8 | 11.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.5 | 5.8 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.4 | 18.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.3 | 5.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 1.3 | 3.9 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 1.2 | 3.6 | GO:1903445 | protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 1.1 | 4.5 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 1.0 | 7.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.9 | 5.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.9 | 2.6 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.7 | 2.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.7 | 2.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.7 | 8.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.5 | 2.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.5 | 3.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.5 | 1.4 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.5 | 0.9 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.4 | 1.8 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.4 | 5.6 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.4 | 3.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 8.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.4 | 1.8 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.3 | 1.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 2.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 10.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.2 | 0.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.2 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 7.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 1.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 6.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 2.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 7.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 3.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 6.7 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 1.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 2.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 7.7 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.1 | 1.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 2.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 4.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 5.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 9.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 5.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 8.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 | 3.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 7.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 2.3 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 2.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 3.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 2.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 8.5 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 1.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.6 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 3.1 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 2.3 | 11.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.8 | 2.3 | GO:0000802 | transverse filament(GO:0000802) |
| 0.8 | 4.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.6 | 5.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.5 | 2.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.4 | 2.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 1.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.3 | 8.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 4.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 2.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 8.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 4.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 12.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 7.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 0.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 7.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 17.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 12.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 5.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 15.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 5.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 17.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.6 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 3.1 | 12.5 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 1.9 | 5.8 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 1.7 | 8.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.4 | 8.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.1 | 11.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 1.1 | 3.4 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.9 | 7.7 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.7 | 4.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.5 | 5.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.5 | 9.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 4.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.4 | 1.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.4 | 11.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.4 | 5.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.4 | 5.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 6.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 11.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 0.9 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 10.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 8.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 2.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 4.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 8.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 3.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 2.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 2.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 4.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 3.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 3.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 3.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 11.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 7.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 11.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.6 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 13.5 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 3.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 5.3 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.2 | 2.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 3.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 2.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 5.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 4.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |