Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Junb_Jund
Z-value: 2.55
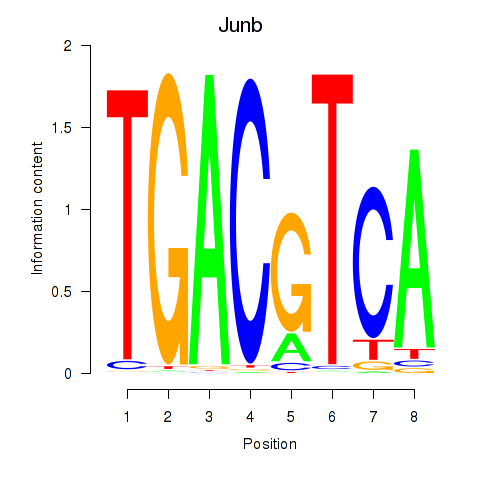
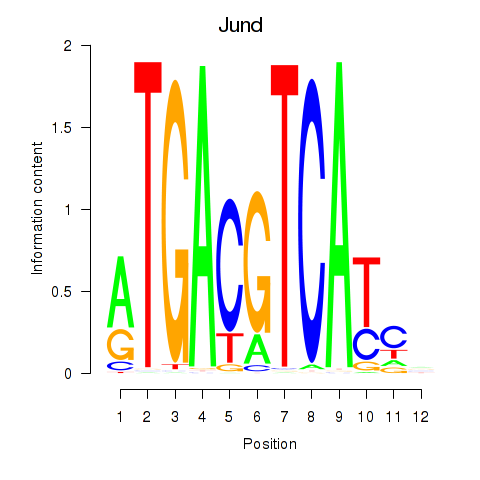
Transcription factors associated with Junb_Jund
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Junb
|
ENSRNOG00000042838 | JunB proto-oncogene, AP-1 transcription factor subunit |
|
Jund
|
ENSRNOG00000019568 | JunD proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Junb | rn6_v1_chr19_-_26094756_26094756 | -0.36 | 3.3e-11 | Click! |
| Jund | rn6_v1_chr16_-_20486707_20486707 | 0.00 | 9.6e-01 | Click! |
Activity profile of Junb_Jund motif
Sorted Z-values of Junb_Jund motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_150801289 | 240.28 |
ENSRNOT00000035060
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr15_+_43007908 | 171.05 |
ENSRNOT00000084753
ENSRNOT00000091567 ENSRNOT00000087709 |
Stmn4
|
stathmin 4 |
| chr8_-_115263484 | 152.65 |
ENSRNOT00000039696
ENSRNOT00000081138 |
Iqcf3
|
IQ motif containing F3 |
| chr6_+_28515025 | 152.46 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr4_+_174181644 | 147.00 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr5_-_146446227 | 146.93 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr5_+_59452348 | 144.25 |
ENSRNOT00000019887
|
Ccin
|
calicin |
| chr2_+_164549455 | 142.83 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr1_-_169334093 | 141.94 |
ENSRNOT00000032587
|
Ubqln3
|
ubiquilin 3 |
| chr5_-_140024176 | 137.98 |
ENSRNOT00000016492
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr15_+_28319136 | 137.03 |
ENSRNOT00000048723
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr4_-_169036950 | 136.44 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr15_+_80040842 | 133.26 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr1_-_89194602 | 130.22 |
ENSRNOT00000028518
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr4_+_159403501 | 129.73 |
ENSRNOT00000086525
|
Akap3
|
A-kinase anchoring protein 3 |
| chr8_-_115274165 | 129.40 |
ENSRNOT00000056386
|
LOC102550160
|
IQ domain-containing protein F5-like |
| chr7_-_124982566 | 128.97 |
ENSRNOT00000075099
|
Sult4a1
|
sulfotransferase family 4A, member 1 |
| chr7_+_18310624 | 122.81 |
ENSRNOT00000075258
|
Actl9
|
actin-like 9 |
| chr11_+_88424414 | 122.15 |
ENSRNOT00000022328
|
Spag6l
|
sperm associated antigen 6-like |
| chr6_+_36089433 | 119.29 |
ENSRNOT00000090438
ENSRNOT00000005731 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr6_-_86223052 | 116.51 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr9_-_85243001 | 113.30 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chrX_+_142344878 | 112.13 |
ENSRNOT00000056621
|
LOC108349258
|
high mobility group protein B4-like |
| chr2_+_104290726 | 110.55 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr16_-_82288022 | 110.23 |
ENSRNOT00000078609
|
Spaca7
|
sperm acrosome associated 7 |
| chr6_-_47848026 | 109.70 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chrX_-_144001727 | 109.59 |
ENSRNOT00000078404
|
RGD1560585
|
similar to RIKEN cDNA 1700001F22 |
| chr2_-_29768750 | 108.93 |
ENSRNOT00000023460
|
Map1b
|
microtubule-associated protein 1B |
| chr9_-_113358526 | 108.33 |
ENSRNOT00000065073
|
Txndc2
|
thioredoxin domain containing 2 |
| chr9_+_93445002 | 107.07 |
ENSRNOT00000025029
|
LOC501180
|
similar to hypothetical protein MGC35154 |
| chr15_-_57651041 | 105.70 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr1_-_89190128 | 104.88 |
ENSRNOT00000067813
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr5_+_73496027 | 104.63 |
ENSRNOT00000022395
|
Actl7a
|
actin-like 7a |
| chr9_-_63291350 | 101.86 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr9_+_10172832 | 101.58 |
ENSRNOT00000074555
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr18_-_14016713 | 100.94 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr1_-_198267093 | 100.14 |
ENSRNOT00000047477
|
RGD1563217
|
similar to RIKEN cDNA 4930451I11 |
| chr1_-_188245628 | 99.66 |
ENSRNOT00000022800
|
Tmc7
|
transmembrane channel-like 7 |
| chr10_+_89918373 | 96.06 |
ENSRNOT00000055192
|
RGD1565533
|
RGD1565533 |
| chr4_-_157078130 | 95.28 |
ENSRNOT00000015570
|
Clstn3
|
calsyntenin 3 |
| chr4_-_6449949 | 94.11 |
ENSRNOT00000062026
|
Galntl5
|
polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr8_+_115213471 | 92.45 |
ENSRNOT00000017570
|
Iqcf5
|
IQ motif containing F5 |
| chr1_-_81295442 | 92.20 |
ENSRNOT00000030900
|
Irgc
|
immunity related GTPase cinema |
| chrX_-_132424746 | 92.12 |
ENSRNOT00000087819
|
LOC681067
|
similar to RIKEN cDNA 1700001F22 |
| chr3_+_170475831 | 91.96 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr8_+_115193146 | 91.14 |
ENSRNOT00000056391
|
Iqcf1
|
IQ motif containing F1 |
| chrX_-_139329975 | 90.67 |
ENSRNOT00000086405
|
LOC103694517
|
high mobility group protein B4-like |
| chrX_-_130794673 | 90.61 |
ENSRNOT00000080612
|
LOC691030
|
similar to RIKEN cDNA 1700001F22 |
| chr1_+_211582077 | 89.90 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr15_-_5467047 | 89.77 |
ENSRNOT00000042743
|
Spetex-2F
|
Spetex-2F protein |
| chrX_+_137197396 | 89.03 |
ENSRNOT00000082227
|
RGD1566225
|
similar to RIKEN cDNA 1700001F22 |
| chr2_+_260039651 | 87.64 |
ENSRNOT00000073873
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chrX_+_114929029 | 86.77 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr1_+_97712177 | 86.50 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr6_+_122603269 | 84.54 |
ENSRNOT00000005297
|
Spata7
|
spermatogenesis associated 7 |
| chr10_+_103992309 | 82.02 |
ENSRNOT00000065292
|
Trim80
|
tripartite motif protein 80 |
| chr5_-_75116490 | 81.79 |
ENSRNOT00000042788
|
Txndc8
|
thioredoxin domain containing 8 |
| chr4_+_56556507 | 81.26 |
ENSRNOT00000060353
|
Fam71f2
|
family with sequence similarity 71, member F2 |
| chr1_-_52495582 | 80.42 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chr17_+_85382116 | 78.93 |
ENSRNOT00000002513
|
Spag6
|
sperm associated antigen 6 |
| chr1_+_101603222 | 78.90 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr18_+_45023932 | 78.48 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr16_-_49574314 | 77.81 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr9_-_100253609 | 77.80 |
ENSRNOT00000036061
|
Kif1a
|
kinesin family member 1A |
| chr3_+_125428260 | 76.97 |
ENSRNOT00000028892
|
Chgb
|
chromogranin B |
| chr1_-_31055453 | 76.74 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chrX_-_158261717 | 75.91 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr15_-_6587367 | 75.34 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr14_-_108284619 | 74.91 |
ENSRNOT00000078814
|
LOC690352
|
hypothetical protein LOC690352 |
| chr6_-_41870046 | 74.70 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr9_-_14906410 | 73.18 |
ENSRNOT00000061674
|
LOC680955
|
hypothetical protein LOC680955 |
| chr14_-_108285008 | 72.78 |
ENSRNOT00000087501
|
LOC690352
|
hypothetical protein LOC690352 |
| chrX_+_82217143 | 72.77 |
ENSRNOT00000086866
|
Cylc1
|
cylicin 1 |
| chr5_-_113880911 | 72.27 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr1_+_100501676 | 71.77 |
ENSRNOT00000043724
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr8_+_4085829 | 71.59 |
ENSRNOT00000031286
|
Actl9b
|
actin-like 9b |
| chr10_+_38104558 | 71.53 |
ENSRNOT00000067809
|
Fstl4
|
follistatin-like 4 |
| chr3_-_45210474 | 71.24 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr12_-_13067341 | 71.03 |
ENSRNOT00000029692
|
Fam220a
|
family with sequence similarity 220, member A |
| chr5_-_57896475 | 70.83 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr14_-_9456990 | 69.07 |
ENSRNOT00000002918
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr1_-_72727112 | 68.41 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr3_-_160922341 | 67.38 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr5_-_25584278 | 66.91 |
ENSRNOT00000090579
ENSRNOT00000090376 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_+_224938801 | 66.81 |
ENSRNOT00000073052
|
1700092M07Rik
|
RIKEN cDNA 1700092M07 gene |
| chr8_-_57255263 | 65.27 |
ENSRNOT00000028972
|
LOC100125362
|
hypothetical protein LOC100125362 |
| chr10_-_56511583 | 62.58 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr11_-_32469566 | 61.97 |
ENSRNOT00000002718
|
LOC498063
|
similar to RIKEN cDNA 4930563D23 |
| chr10_+_49368314 | 61.66 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr1_+_72644333 | 61.48 |
ENSRNOT00000048976
ENSRNOT00000065051 |
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr2_-_251970768 | 61.18 |
ENSRNOT00000020141
|
Wdr63
|
WD repeat domain 63 |
| chr3_+_138974871 | 60.34 |
ENSRNOT00000012524
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chrX_-_29648359 | 59.90 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr6_+_109939345 | 59.85 |
ENSRNOT00000013560
|
Ift43
|
intraflagellar transport 43 |
| chr4_-_184096806 | 58.97 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr11_+_52828116 | 58.56 |
ENSRNOT00000035340
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr10_+_57239993 | 57.59 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr2_+_157453428 | 57.44 |
ENSRNOT00000051494
|
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr16_+_20691978 | 57.26 |
ENSRNOT00000038139
ENSRNOT00000082319 |
Tmem59l
|
transmembrane protein 59-like |
| chr13_+_104284660 | 57.18 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr8_-_87282156 | 56.37 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chr4_-_85915099 | 55.98 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr7_+_40318490 | 55.82 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr15_+_23777856 | 55.67 |
ENSRNOT00000060847
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr6_-_126622532 | 54.95 |
ENSRNOT00000038816
|
Moap1
|
modulator of apoptosis 1 |
| chr10_-_14056169 | 54.66 |
ENSRNOT00000017833
|
Syngr3
|
synaptogyrin 3 |
| chr10_-_91699424 | 54.60 |
ENSRNOT00000004650
|
Lyzl6
|
lysozyme-like 6 |
| chr6_+_147876237 | 54.39 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr2_+_220298245 | 53.68 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr13_-_104284068 | 53.08 |
ENSRNOT00000005407
|
LOC100912365
|
uncharacterized LOC100912365 |
| chr17_-_9695292 | 52.79 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr1_-_220467159 | 52.46 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr10_+_82775691 | 52.25 |
ENSRNOT00000030737
|
Hils1
|
histone linker H1 domain, spermatid-specific 1 |
| chr6_+_126434226 | 52.04 |
ENSRNOT00000090857
|
Chga
|
chromogranin A |
| chr8_+_64364741 | 51.57 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr16_+_83161880 | 51.36 |
ENSRNOT00000080793
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr12_+_13390859 | 51.01 |
ENSRNOT00000080489
|
Spdye4
|
speedy/RINGO cell cycle regulator family member E4 |
| chrX_+_13992064 | 50.80 |
ENSRNOT00000036543
|
LOC100363125
|
rCG42854-like |
| chr9_-_16095007 | 50.58 |
ENSRNOT00000073333
|
AABR07066782.1
|
|
| chr1_+_111849998 | 49.81 |
ENSRNOT00000033811
|
Luzp2
|
leucine zipper protein 2 |
| chr10_-_89918427 | 49.34 |
ENSRNOT00000084217
|
Dusp3
|
dual specificity phosphatase 3 |
| chr15_+_52265557 | 48.89 |
ENSRNOT00000015969
|
Nudt18
|
nudix hydrolase 18 |
| chr4_-_156945126 | 48.50 |
ENSRNOT00000064853
|
RGD1307916
|
LOC362433 |
| chr1_-_198382614 | 48.48 |
ENSRNOT00000055016
|
Asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr1_+_102849889 | 47.80 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr9_+_10535340 | 47.68 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr4_-_88565292 | 47.15 |
ENSRNOT00000008948
|
Lancl2
|
LanC like 2 |
| chr19_-_11669578 | 47.07 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr5_-_144779212 | 47.04 |
ENSRNOT00000016230
|
Ncdn
|
neurochondrin |
| chr4_-_183697531 | 46.76 |
ENSRNOT00000055441
|
Amn1
|
antagonist of mitotic exit network 1 homolog |
| chr10_+_49472460 | 46.49 |
ENSRNOT00000038276
|
Tekt3
|
tektin 3 |
| chr3_-_112371664 | 46.03 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chr7_-_140951071 | 45.94 |
ENSRNOT00000082521
|
Fam186b
|
family with sequence similarity 186, member B |
| chr10_-_17721233 | 45.73 |
ENSRNOT00000031337
|
Smim23
|
small integral membrane protein 23 |
| chr18_+_47613854 | 45.50 |
ENSRNOT00000025270
|
Zfp474
|
zinc finger protein 474 |
| chr3_+_121128368 | 44.26 |
ENSRNOT00000055882
|
LOC499886
|
similar to hypothetical protein 4933411G11 |
| chr9_-_93735636 | 43.98 |
ENSRNOT00000025582
|
Nppc
|
natriuretic peptide C |
| chr2_-_104461863 | 43.65 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr8_-_61519507 | 43.60 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr11_+_80358211 | 43.20 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr20_-_37700937 | 43.08 |
ENSRNOT00000001051
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr17_+_1679627 | 42.92 |
ENSRNOT00000025801
|
Habp4
|
hyaluronan binding protein 4 |
| chr3_+_137618898 | 42.84 |
ENSRNOT00000007249
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chrX_+_151103576 | 42.41 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr20_+_29655226 | 42.25 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr11_-_69355854 | 42.15 |
ENSRNOT00000002975
|
Ropn1
|
rhophilin associated tail protein 1 |
| chr5_-_39611053 | 42.14 |
ENSRNOT00000046595
|
Fhl5
|
four and a half LIM domains 5 |
| chr1_-_215033460 | 42.14 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr8_+_58431407 | 41.84 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr20_+_13498926 | 41.40 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr8_-_17524839 | 41.26 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr13_+_49003803 | 41.03 |
ENSRNOT00000080556
|
Lemd1
|
LEM domain containing 1 |
| chr15_-_5417570 | 40.87 |
ENSRNOT00000061525
|
Spetex-2F
|
Spetex-2F protein |
| chr18_-_24057917 | 40.82 |
ENSRNOT00000023874
|
Rit2
|
Ras-like without CAAX 2 |
| chr10_+_75620470 | 40.35 |
ENSRNOT00000013882
|
Ccdc182
|
coiled-coil domain containing 182 |
| chr1_+_97777685 | 38.57 |
ENSRNOT00000030895
|
LOC690284
|
similar to F49E2.5d |
| chr2_-_30748325 | 38.40 |
ENSRNOT00000084294
ENSRNOT00000083089 |
Mrps36
|
mitochondrial ribosomal protein S36 |
| chr9_-_19613360 | 38.29 |
ENSRNOT00000029593
|
Rcan2
|
regulator of calcineurin 2 |
| chr10_-_91186054 | 38.27 |
ENSRNOT00000004140
|
Plcd3
|
phospholipase C, delta 3 |
| chr8_+_48422036 | 37.80 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr8_+_60117729 | 37.61 |
ENSRNOT00000021074
|
Isl2
|
ISL LIM homeobox 2 |
| chr9_-_28732919 | 37.55 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr5_-_65073012 | 37.18 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr9_+_102862890 | 37.05 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr13_+_88644520 | 37.02 |
ENSRNOT00000003979
|
Spata46
|
spermatogenesis associated 46 |
| chr10_+_94192556 | 36.52 |
ENSRNOT00000074007
ENSRNOT00000093039 |
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr9_+_82674202 | 36.49 |
ENSRNOT00000027208
|
Tmem198
|
transmembrane protein 198 |
| chr17_+_27452638 | 36.38 |
ENSRNOT00000038349
|
Cage1
|
cancer antigen 1 |
| chr1_+_218945508 | 36.02 |
ENSRNOT00000034240
|
RGD1311946
|
similar to RIKEN cDNA 1810055G02 |
| chr4_+_70977556 | 35.09 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr7_-_72328128 | 34.70 |
ENSRNOT00000008227
|
Tspyl5
|
TSPY-like 5 |
| chr5_+_103479767 | 34.66 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr10_+_88356615 | 34.64 |
ENSRNOT00000022687
|
Klhl10
|
kelch-like family member 10 |
| chr11_-_87240833 | 34.44 |
ENSRNOT00000052200
|
Tssk1b
|
testis-specific serine kinase 1B |
| chr20_-_12835044 | 34.43 |
ENSRNOT00000074268
|
LOC108348157
|
speriolin-like protein |
| chr2_+_233602732 | 34.28 |
ENSRNOT00000044232
|
Pitx2
|
paired-like homeodomain 2 |
| chr1_+_211205903 | 34.16 |
ENSRNOT00000023139
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr6_+_98284170 | 33.95 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr1_-_225035687 | 33.91 |
ENSRNOT00000026539
|
Gng3
|
G protein subunit gamma 3 |
| chr10_-_36716601 | 33.83 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr9_+_82053581 | 33.81 |
ENSRNOT00000086375
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
| chr1_+_201913148 | 33.64 |
ENSRNOT00000036128
|
Fam24a
|
family with sequence similarity 24, member A |
| chr14_+_8080275 | 33.31 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr9_+_99998275 | 32.99 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr20_-_45815940 | 32.52 |
ENSRNOT00000073276
|
Gpr6
|
G protein-coupled receptor 6 |
| chr1_-_24604400 | 32.43 |
ENSRNOT00000081175
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr7_+_123168811 | 32.43 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr6_-_42473738 | 32.32 |
ENSRNOT00000033327
|
Kcnf1
|
potassium voltage-gated channel modifier subfamily F member 1 |
| chr3_-_3266260 | 31.88 |
ENSRNOT00000034538
|
Glt6d1
|
glycosyltransferase 6 domain containing 1 |
| chrX_-_82986051 | 31.84 |
ENSRNOT00000077587
|
Hdx
|
highly divergent homeobox |
| chr15_-_47442664 | 31.47 |
ENSRNOT00000072994
|
Prss55
|
protease, serine, 55 |
| chr3_+_150910398 | 31.42 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr7_-_44121130 | 31.29 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr6_-_110681408 | 31.25 |
ENSRNOT00000078265
|
Angel1
|
angel homolog 1 |
| chr10_+_70520206 | 31.23 |
ENSRNOT00000088198
ENSRNOT00000090446 ENSRNOT00000085799 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr17_+_7675531 | 31.21 |
ENSRNOT00000061187
|
Spock1
|
sparc/osteonectin, cwcv and kazal like domains proteoglycan 1 |
| chr7_+_72924799 | 30.96 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr10_+_89376530 | 30.92 |
ENSRNOT00000028089
|
Rnd2
|
Rho family GTPase 2 |
| chr10_-_82252720 | 30.72 |
ENSRNOT00000066132
ENSRNOT00000075795 |
Mycbpap
|
Mycbp associated protein |
| chr10_-_27179900 | 30.62 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Junb_Jund
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 36.6 | 109.7 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 22.8 | 91.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 20.4 | 265.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 18.8 | 37.6 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 17.3 | 52.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 16.4 | 49.3 | GO:0060577 | pulmonary vein morphogenesis(GO:0060577) |
| 16.3 | 48.9 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 16.0 | 64.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 16.0 | 240.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 15.7 | 47.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 15.6 | 77.8 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 15.5 | 46.5 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 14.4 | 129.4 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 14.3 | 57.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 12.5 | 37.6 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 11.1 | 111.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 11.0 | 33.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 11.0 | 44.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 11.0 | 55.0 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 10.8 | 183.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 10.7 | 74.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 10.6 | 84.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 9.6 | 38.6 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 9.6 | 105.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 9.6 | 77.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 9.5 | 95.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 9.4 | 18.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 9.1 | 18.2 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 8.9 | 26.7 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 8.7 | 61.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 8.7 | 26.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 8.6 | 42.8 | GO:0030070 | insulin processing(GO:0030070) |
| 8.4 | 108.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 8.1 | 122.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 8.1 | 105.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 8.0 | 23.9 | GO:1900222 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 7.9 | 71.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 7.8 | 31.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 7.6 | 113.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 7.0 | 21.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 6.8 | 40.8 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 6.7 | 33.4 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 6.6 | 26.5 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 6.6 | 26.3 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 6.3 | 50.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 6.3 | 31.3 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 6.0 | 59.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 5.9 | 147.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 5.8 | 34.6 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 5.4 | 16.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 5.3 | 42.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 5.3 | 5.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 5.2 | 36.3 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) |
| 5.1 | 45.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 5.1 | 25.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 5.0 | 55.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 5.0 | 34.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 4.5 | 9.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 4.5 | 13.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 4.5 | 17.9 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 4.4 | 13.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 4.4 | 17.4 | GO:0015888 | thiamine transport(GO:0015888) |
| 4.3 | 17.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 4.3 | 21.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 4.1 | 12.2 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 4.0 | 12.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 3.8 | 19.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 3.8 | 34.2 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 3.8 | 7.6 | GO:0019086 | late viral transcription(GO:0019086) |
| 3.7 | 14.8 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 3.7 | 88.6 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 3.6 | 277.3 | GO:0030317 | sperm motility(GO:0030317) |
| 3.6 | 43.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 3.6 | 25.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 3.6 | 21.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 3.5 | 17.6 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 3.5 | 145.8 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 3.4 | 10.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 3.3 | 29.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 3.2 | 38.2 | GO:0007614 | short-term memory(GO:0007614) |
| 3.2 | 41.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 3.2 | 25.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 3.1 | 56.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 3.1 | 25.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 3.1 | 24.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 3.0 | 63.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 3.0 | 74.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 2.9 | 14.7 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 2.9 | 31.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 2.9 | 11.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 2.9 | 8.6 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 2.8 | 11.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.8 | 59.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 2.8 | 30.6 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 2.7 | 10.6 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 2.7 | 13.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 2.6 | 36.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 2.5 | 17.7 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 2.5 | 10.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 2.5 | 19.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 2.4 | 9.7 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 2.4 | 37.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 2.4 | 7.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 2.3 | 84.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 2.3 | 33.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 2.2 | 22.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 2.2 | 55.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 2.2 | 17.8 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 2.2 | 19.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 2.2 | 13.0 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 2.2 | 8.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 2.2 | 13.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 2.0 | 24.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 2.0 | 13.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 1.9 | 9.7 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 1.9 | 27.1 | GO:0042026 | protein refolding(GO:0042026) |
| 1.9 | 11.6 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 1.9 | 19.0 | GO:0032782 | bile acid secretion(GO:0032782) |
| 1.9 | 5.6 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 1.9 | 24.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 1.8 | 9.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.8 | 5.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.7 | 42.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.7 | 139.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 1.7 | 5.2 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 1.7 | 15.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.7 | 8.6 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 1.7 | 6.8 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.7 | 43.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 1.6 | 52.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 1.6 | 30.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.6 | 17.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.6 | 6.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 1.6 | 19.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 1.6 | 40.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 1.5 | 39.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 1.5 | 6.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 1.5 | 6.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.5 | 4.5 | GO:1990792 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 1.5 | 5.9 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 1.4 | 10.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 1.4 | 18.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.4 | 8.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.4 | 17.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 1.3 | 26.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 1.3 | 41.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 1.3 | 12.9 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 1.3 | 9.0 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.3 | 35.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 1.3 | 3.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 1.2 | 14.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 1.2 | 7.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 1.2 | 19.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 1.2 | 176.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 1.2 | 113.1 | GO:0007338 | single fertilization(GO:0007338) |
| 1.2 | 1.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.2 | 147.4 | GO:0007286 | spermatid development(GO:0007286) |
| 1.2 | 19.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 1.1 | 13.6 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 1.1 | 26.9 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 1.1 | 28.6 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 1.1 | 9.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 1.1 | 54.3 | GO:0021762 | substantia nigra development(GO:0021762) |
| 1.1 | 24.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 1.0 | 8.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 1.0 | 3.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.0 | 22.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 1.0 | 10.8 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 1.0 | 14.6 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 1.0 | 5.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 1.0 | 101.6 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 1.0 | 60.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.9 | 14.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.9 | 10.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.9 | 11.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.9 | 34.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.8 | 47.1 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.8 | 9.8 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.8 | 26.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.7 | 3.7 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.7 | 10.8 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.7 | 7.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.7 | 15.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.7 | 27.6 | GO:0009994 | oocyte differentiation(GO:0009994) |
| 0.7 | 4.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.7 | 42.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.7 | 8.0 | GO:0021924 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.6 | 15.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.6 | 3.0 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.6 | 9.4 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.6 | 4.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.6 | 1.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.6 | 1.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 3.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.5 | 2.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.5 | 13.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.5 | 9.0 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.5 | 30.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.5 | 29.1 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.5 | 2.4 | GO:0015819 | lysine transport(GO:0015819) L-lysine transport(GO:1902022) L-lysine transmembrane transport(GO:1903401) |
| 0.5 | 3.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.5 | 8.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.4 | 36.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.4 | 1.3 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.4 | 3.9 | GO:1904874 | regulation of telomerase RNA localization to Cajal body(GO:1904872) positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.4 | 51.5 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.4 | 16.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.4 | 12.6 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.4 | 7.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.4 | 3.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.4 | 41.9 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.4 | 6.3 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.4 | 11.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.4 | 6.0 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.4 | 2.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.4 | 31.0 | GO:0008585 | female gonad development(GO:0008585) |
| 0.4 | 99.5 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.4 | 2.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 5.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 10.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.4 | 9.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.3 | 0.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.3 | 2.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 2.6 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.3 | 18.8 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.3 | 1.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.2 | 2.7 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.2 | 1.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 35.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.2 | 32.0 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.2 | 14.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.2 | 2.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 4.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 1.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 32.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 0.8 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 1.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 6.0 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 11.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 1.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.7 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 9.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.4 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.1 | 0.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 1.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 2.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 3.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 4.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 1.4 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 3.3 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 2.0 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.1 | 529.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 24.4 | 122.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 18.1 | 72.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 18.0 | 90.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 16.0 | 240.3 | GO:0044754 | autolysosome(GO:0044754) |
| 14.7 | 147.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 10.5 | 42.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 9.6 | 38.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 9.4 | 66.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 8.6 | 59.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 8.3 | 41.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 8.0 | 160.2 | GO:0043196 | varicosity(GO:0043196) |
| 7.1 | 70.8 | GO:0002177 | manchette(GO:0002177) |
| 6.9 | 103.2 | GO:0031045 | dense core granule(GO:0031045) |
| 6.5 | 156.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 6.5 | 52.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 6.5 | 19.4 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 6.0 | 108.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 5.9 | 17.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 5.7 | 22.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 5.5 | 125.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 5.3 | 47.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 4.7 | 37.6 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 4.3 | 29.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 4.1 | 423.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 4.0 | 12.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 4.0 | 24.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 3.9 | 55.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 3.6 | 28.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 3.3 | 13.2 | GO:0097196 | Shu complex(GO:0097196) |
| 2.6 | 18.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 2.5 | 19.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 2.4 | 35.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 2.2 | 37.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 2.2 | 6.5 | GO:0035577 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 2.1 | 4.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 2.1 | 44.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 2.1 | 59.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 2.0 | 81.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 2.0 | 147.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 1.9 | 5.8 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 1.9 | 70.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 1.8 | 49.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.8 | 180.4 | GO:0031514 | motile cilium(GO:0031514) |
| 1.8 | 5.4 | GO:0044308 | axonal spine(GO:0044308) |
| 1.8 | 21.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.7 | 12.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.6 | 14.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.6 | 9.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 1.5 | 6.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.5 | 9.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.5 | 16.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 1.5 | 14.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.4 | 26.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.3 | 25.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 1.3 | 30.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.3 | 31.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 1.3 | 6.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.2 | 33.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 1.2 | 7.2 | GO:0098871 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 1.1 | 28.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 1.1 | 30.2 | GO:0031430 | M band(GO:0031430) |
| 1.1 | 4.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.1 | 11.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 1.1 | 182.4 | GO:0030426 | growth cone(GO:0030426) |
| 1.0 | 231.5 | GO:0030141 | secretory granule(GO:0030141) |
| 1.0 | 131.5 | GO:0043204 | perikaryon(GO:0043204) |
| 1.0 | 33.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.0 | 12.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.0 | 88.4 | GO:0030018 | Z disc(GO:0030018) |
| 1.0 | 8.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.9 | 46.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.9 | 185.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.8 | 6.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.8 | 9.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.8 | 9.7 | GO:0000801 | central element(GO:0000801) |
| 0.8 | 23.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.7 | 26.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.7 | 10.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.7 | 12.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.6 | 5.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.6 | 24.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.6 | 6.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.6 | 10.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.6 | 115.4 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.5 | 15.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.5 | 20.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.5 | 9.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.5 | 32.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.5 | 27.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.5 | 57.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.5 | 11.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 53.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.4 | 103.8 | GO:0005768 | endosome(GO:0005768) |
| 0.4 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 4.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.4 | 6.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) AMPA glutamate receptor complex(GO:0032281) neurotransmitter receptor complex(GO:0098878) |
| 0.4 | 23.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.4 | 8.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 40.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.3 | 8.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 89.9 | GO:0030424 | axon(GO:0030424) |
| 0.3 | 15.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 4.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 3.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 0.8 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.3 | 13.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 69.3 | GO:0045202 | synapse(GO:0045202) |
| 0.2 | 2.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 0.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 11.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 23.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 3.9 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 2.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 7.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 6.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 5.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 5.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 9.0 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 24.0 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 3.0 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 5.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 1.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 35.1 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 2.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 8.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 125.2 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.6 | GO:0045177 | apical part of cell(GO:0045177) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 58.8 | 235.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 24.0 | 240.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 23.8 | 190.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 23.7 | 71.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 23.0 | 69.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 22.3 | 66.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 19.7 | 59.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 16.4 | 98.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 16.3 | 48.9 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 14.3 | 71.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 12.6 | 50.4 | GO:0030614 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 11.5 | 80.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 10.2 | 30.5 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 9.6 | 162.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 9.1 | 36.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 8.8 | 78.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 8.5 | 101.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 8.4 | 108.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 7.3 | 123.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 7.2 | 86.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 6.6 | 33.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 6.3 | 25.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 6.2 | 43.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 5.8 | 17.4 | GO:0004040 | amidase activity(GO:0004040) |
| 5.7 | 22.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 5.6 | 22.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 5.5 | 55.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 5.4 | 70.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 5.4 | 26.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 5.1 | 25.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 5.0 | 89.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 4.6 | 27.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 4.4 | 17.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 4.2 | 54.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 4.1 | 41.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 4.1 | 69.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 4.0 | 113.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 3.9 | 73.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 3.8 | 11.5 | GO:0017098 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 3.7 | 37.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 3.6 | 17.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 3.3 | 33.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 3.2 | 9.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 3.1 | 40.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 3.0 | 6.0 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 3.0 | 149.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 3.0 | 21.0 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 3.0 | 76.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 2.9 | 5.8 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 2.8 | 73.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 2.8 | 19.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 2.8 | 49.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 2.7 | 24.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 2.7 | 56.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 2.7 | 47.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 2.7 | 8.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 2.6 | 31.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 2.6 | 31.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 2.5 | 10.0 | GO:0016492 | nerve growth factor receptor activity(GO:0010465) G-protein coupled neurotensin receptor activity(GO:0016492) |
| 2.4 | 31.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 2.4 | 72.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 2.4 | 23.9 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 2.3 | 52.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 2.3 | 419.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 2.2 | 11.0 | GO:0080084 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) 5S rDNA binding(GO:0080084) |
| 2.2 | 30.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 2.2 | 13.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 2.1 | 14.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 2.1 | 129.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 2.0 | 6.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 2.0 | 6.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 2.0 | 38.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 1.9 | 52.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 1.9 | 25.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.9 | 19.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.9 | 15.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.9 | 5.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 1.8 | 16.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.8 | 17.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 1.8 | 5.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.7 | 42.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.7 | 95.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 1.5 | 24.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.5 | 25.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.5 | 37.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 1.4 | 13.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 1.4 | 8.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.4 | 17.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.3 | 18.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 1.2 | 17.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 1.2 | 6.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.2 | 10.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.2 | 12.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 1.1 | 14.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 1.1 | 4.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.1 | 18.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 1.1 | 52.9 | GO:0042805 | actinin binding(GO:0042805) |
| 1.0 | 3.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.0 | 319.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 1.0 | 5.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 1.0 | 3.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.0 | 5.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.0 | 50.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 1.0 | 6.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 1.0 | 28.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 1.0 | 18.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.9 | 35.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.9 | 10.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.9 | 89.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.9 | 2.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.9 | 5.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.9 | 10.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.9 | 8.6 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.9 | 63.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.8 | 13.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.8 | 23.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 22.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.7 | 81.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.7 | 60.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.7 | 25.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.7 | 9.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 22.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.7 | 12.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.7 | 33.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.6 | 7.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.6 | 41.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.6 | 5.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.6 | 19.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.5 | 14.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 8.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.4 | 111.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.4 | 12.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.4 | 14.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 1.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.4 | 2.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 11.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 11.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 32.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.4 | 10.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 1.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.3 | 9.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.3 | 71.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 16.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.3 | 6.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 32.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 71.6 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.3 | 36.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 23.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.2 | 8.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 79.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.2 | 2.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 1.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 3.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 2.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 4.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 3.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 9.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 3.8 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 2.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 32.3 | GO:0008289 | lipid binding(GO:0008289) |
| 0.1 | 4.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 4.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 38.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 3.0 | 44.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 2.6 | 109.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 2.5 | 68.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 2.4 | 25.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 2.2 | 85.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 1.9 | 67.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 1.4 | 21.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.9 | 8.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.9 | 38.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.8 | 38.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.8 | 21.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.7 | 6.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.6 | 4.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.6 | 8.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.6 | 22.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.5 | 14.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.5 | 10.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.5 | 8.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.4 | 8.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 14.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 16.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.3 | 2.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 5.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 3.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 11.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 119.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 9.9 | 129.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 7.0 | 274.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 6.3 | 56.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 4.5 | 66.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 4.2 | 105.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 3.9 | 65.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 3.4 | 47.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 3.2 | 13.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 3.0 | 17.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 2.8 | 41.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 2.7 | 8.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 2.7 | 81.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 2.4 | 55.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 2.3 | 37.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 2.2 | 47.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 1.9 | 53.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 1.8 | 39.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.8 | 30.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.8 | 33.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 1.7 | 50.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 1.7 | 18.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 1.6 | 28.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 1.6 | 35.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 1.5 | 20.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.4 | 72.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 1.4 | 26.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 1.3 | 13.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.3 | 18.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 1.2 | 13.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 1.2 | 24.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 1.1 | 17.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 1.0 | 84.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 1.0 | 24.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.9 | 16.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.8 | 24.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.8 | 30.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.7 | 38.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.7 | 21.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 56.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.6 | 11.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.6 | 4.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.6 | 8.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.6 | 9.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.6 | 18.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.5 | 6.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 6.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 24.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 6.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 3.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 14.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 6.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 8.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 2.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 8.6 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.2 | 10.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 2.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 7.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 11.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |