Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
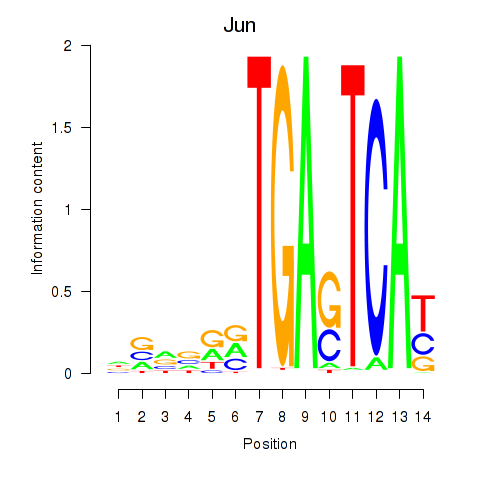
Results for Jun
Z-value: 1.02
Transcription factors associated with Jun
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Jun
|
ENSRNOG00000026293 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Jun | rn6_v1_chr5_-_114014277_114014277 | -0.07 | 1.9e-01 | Click! |
Activity profile of Jun motif
Sorted Z-values of Jun motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_199555722 | 73.26 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr4_+_98457810 | 63.87 |
ENSRNOT00000074175
|
AABR07060872.1
|
|
| chr12_+_24761210 | 41.43 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr8_+_5606592 | 40.72 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr13_+_27889345 | 37.67 |
ENSRNOT00000079134
ENSRNOT00000003274 |
Serpinb8
|
serpin family B member 8 |
| chr4_+_78694447 | 35.78 |
ENSRNOT00000011945
|
Gpnmb
|
glycoprotein nmb |
| chr10_+_83655460 | 30.12 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr15_-_35417273 | 28.38 |
ENSRNOT00000083961
ENSRNOT00000041430 |
Gzmb
|
granzyme B |
| chr8_+_55603968 | 28.27 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr3_+_16753703 | 28.25 |
ENSRNOT00000077741
|
AABR07051548.2
|
|
| chr1_-_197770669 | 27.15 |
ENSRNOT00000023563
|
Lat
|
linker for activation of T cells |
| chr13_+_88606894 | 26.17 |
ENSRNOT00000048692
|
Sh2d1b
|
SH2 domain containing 1B |
| chr4_+_99239115 | 25.36 |
ENSRNOT00000009515
|
Cd8a
|
CD8a molecule |
| chr4_-_164049599 | 25.24 |
ENSRNOT00000078267
|
AABR07062183.1
|
|
| chr7_-_142300382 | 25.23 |
ENSRNOT00000048262
|
Bin2
|
bridging integrator 2 |
| chr20_+_295250 | 24.99 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr1_+_220992770 | 24.99 |
ENSRNOT00000045233
|
Rela
|
RELA proto-oncogene, NF-kB subunit |
| chr10_-_15125408 | 24.62 |
ENSRNOT00000026395
|
Msln
|
mesothelin |
| chr3_+_19274273 | 24.09 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chr4_-_163445302 | 23.91 |
ENSRNOT00000087106
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr15_-_36472327 | 23.41 |
ENSRNOT00000032601
ENSRNOT00000059660 |
LOC102553861
|
granzyme-like protein 1-like |
| chr13_+_92264231 | 22.81 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr3_+_19772056 | 22.66 |
ENSRNOT00000044455
|
AABR07051708.1
|
|
| chr3_+_18315320 | 22.61 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr1_+_81499821 | 22.41 |
ENSRNOT00000027100
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr13_+_89975267 | 21.50 |
ENSRNOT00000006266
ENSRNOT00000000053 |
Cd244
|
CD244 molecule |
| chr4_-_163423628 | 21.31 |
ENSRNOT00000079526
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr1_-_89474252 | 21.10 |
ENSRNOT00000028597
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr3_-_14019204 | 21.01 |
ENSRNOT00000072400
ENSRNOT00000092918 |
Traf1
|
TNF receptor-associated factor 1 |
| chr4_-_155401480 | 21.00 |
ENSRNOT00000020735
|
Apobec1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr11_+_27364916 | 20.90 |
ENSRNOT00000002151
|
Bach1
|
BTB domain and CNC homolog 1 |
| chr1_-_99587205 | 20.89 |
ENSRNOT00000048947
|
Siglec8
|
sialic acid binding Ig-like lectin 8 |
| chr18_-_28535828 | 20.72 |
ENSRNOT00000068386
|
Tmem173
|
transmembrane protein 173 |
| chrX_+_28593405 | 19.78 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr9_+_67774150 | 18.91 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr18_-_28438654 | 18.85 |
ENSRNOT00000036301
|
Mzb1
|
marginal zone B and B1 cell-specific protein |
| chr1_-_89473904 | 18.55 |
ENSRNOT00000089474
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr3_-_16753987 | 18.52 |
ENSRNOT00000091257
|
AABR07051548.1
|
|
| chr3_+_162346490 | 18.25 |
ENSRNOT00000087530
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr20_+_5040337 | 17.96 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr3_+_16610086 | 17.77 |
ENSRNOT00000046231
|
LOC100361009
|
rCG64257-like |
| chr1_+_140601791 | 17.33 |
ENSRNOT00000091588
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr4_-_163261958 | 17.20 |
ENSRNOT00000086390
|
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr2_+_248249468 | 16.96 |
ENSRNOT00000022648
|
Gbp4
|
guanylate binding protein 4 |
| chr3_-_166994286 | 16.40 |
ENSRNOT00000081593
|
Zfp217
|
zinc finger protein 217 |
| chr3_-_166993940 | 16.37 |
ENSRNOT00000034669
|
Zfp217
|
zinc finger protein 217 |
| chr17_+_10138810 | 16.23 |
ENSRNOT00000079850
|
Hk3
|
hexokinase 3 |
| chr8_-_128665988 | 16.18 |
ENSRNOT00000050543
|
Csrnp1
|
cysteine and serine rich nuclear protein 1 |
| chr13_-_70626252 | 16.04 |
ENSRNOT00000036947
|
Lamc2
|
laminin subunit gamma 2 |
| chr4_-_163445136 | 15.98 |
ENSRNOT00000080299
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr11_+_61970976 | 15.52 |
ENSRNOT00000078921
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr15_+_36897999 | 15.37 |
ENSRNOT00000076030
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chrX_+_134940615 | 15.20 |
ENSRNOT00000005604
|
Xpnpep2
|
X-prolyl aminopeptidase 2 |
| chr16_+_9235888 | 15.06 |
ENSRNOT00000027204
|
Lrrc18
|
leucine rich repeat containing 18 |
| chr6_-_11298216 | 14.87 |
ENSRNOT00000021135
|
Epcam
|
epithelial cell adhesion molecule |
| chr13_-_36101411 | 14.80 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr15_+_36898195 | 14.69 |
ENSRNOT00000076266
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr3_+_151285249 | 14.43 |
ENSRNOT00000055254
|
Procr
|
protein C receptor |
| chr14_+_89314176 | 14.39 |
ENSRNOT00000006765
|
Upp1
|
uridine phosphorylase 1 |
| chr2_+_248276709 | 14.36 |
ENSRNOT00000068683
|
Gbp2
|
guanylate binding protein 2 |
| chr2_-_26135340 | 14.27 |
ENSRNOT00000074626
|
F2r
|
coagulation factor II (thrombin) receptor |
| chr1_+_81230612 | 14.07 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr3_-_92749121 | 14.03 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chr1_+_221236773 | 13.83 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr4_+_71740532 | 13.72 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr7_+_145068286 | 13.57 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr1_+_81230989 | 13.50 |
ENSRNOT00000077952
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr1_-_78180216 | 13.48 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr1_-_191007503 | 13.26 |
ENSRNOT00000023262
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr2_-_60657712 | 13.21 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr20_+_3189473 | 13.17 |
ENSRNOT00000047439
|
RT1-T24-4
|
RT1 class I, locus T24, gene 4 |
| chr6_-_138852571 | 13.14 |
ENSRNOT00000081803
|
AABR07065656.8
|
|
| chr4_-_103761881 | 13.13 |
ENSRNOT00000084103
|
AABR07061087.1
|
|
| chr13_+_26903052 | 12.69 |
ENSRNOT00000003625
|
Serpinb5
|
serpin family B member 5 |
| chr5_-_159293673 | 12.63 |
ENSRNOT00000009081
|
Padi4
|
peptidyl arginine deiminase 4 |
| chr3_+_164665532 | 12.59 |
ENSRNOT00000014309
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr4_+_96449351 | 12.42 |
ENSRNOT00000091272
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr4_-_22307453 | 12.03 |
ENSRNOT00000047126
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr1_-_228194977 | 11.73 |
ENSRNOT00000028535
|
Stx3
|
syntaxin 3 |
| chr4_-_10329241 | 11.68 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr18_+_56414488 | 11.62 |
ENSRNOT00000088988
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr19_+_55381565 | 11.62 |
ENSRNOT00000018923
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr4_+_3043231 | 11.49 |
ENSRNOT00000013732
|
Il6
|
interleukin 6 |
| chr3_+_16590244 | 11.18 |
ENSRNOT00000073229
|
AABR07051535.1
|
|
| chr10_-_88152064 | 10.66 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chr5_-_151824633 | 10.30 |
ENSRNOT00000043959
|
Sfn
|
stratifin |
| chr11_-_87628502 | 10.22 |
ENSRNOT00000002568
|
Med15
|
mediator complex subunit 15 |
| chr10_+_105499569 | 10.01 |
ENSRNOT00000088457
|
Sphk1
|
sphingosine kinase 1 |
| chr5_-_163167299 | 9.96 |
ENSRNOT00000022478
|
Tnfrsf1b
|
TNF receptor superfamily member 1B |
| chr8_+_117117430 | 9.87 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr13_-_70625842 | 9.69 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr7_-_11648322 | 9.62 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr3_+_161413298 | 9.53 |
ENSRNOT00000023965
ENSRNOT00000088776 |
Mmp9
|
matrix metallopeptidase 9 |
| chr10_-_83655182 | 9.51 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr10_+_47961056 | 9.46 |
ENSRNOT00000027312
|
Fam83g
|
family with sequence similarity 83, member G |
| chr3_+_55623634 | 9.34 |
ENSRNOT00000080525
|
Dhrs9
|
dehydrogenase/reductase 9 |
| chr10_-_88036040 | 9.28 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chrX_+_43625169 | 9.20 |
ENSRNOT00000086311
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr16_-_10706073 | 9.07 |
ENSRNOT00000089114
|
Fam25a
|
family with sequence similarity 25, member A |
| chr6_-_1454480 | 8.95 |
ENSRNOT00000072810
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_+_1452644 | 8.89 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr4_+_169147243 | 8.72 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr8_+_75687100 | 8.64 |
ENSRNOT00000038677
|
Anxa2
|
annexin A2 |
| chrX_+_104734082 | 8.58 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr4_+_169161585 | 8.54 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr15_+_87704340 | 8.52 |
ENSRNOT00000034987
|
Scel
|
sciellin |
| chr4_-_131694755 | 8.46 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr3_-_119619865 | 8.38 |
ENSRNOT00000016172
|
Itpripl1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr10_+_15485905 | 8.34 |
ENSRNOT00000027609
|
Tmem8a
|
transmembrane protein 8A |
| chr18_+_3861539 | 8.32 |
ENSRNOT00000015363
|
Lama3
|
laminin subunit alpha 3 |
| chr20_+_10438444 | 8.28 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr12_-_51877624 | 8.18 |
ENSRNOT00000056800
|
Chek2
|
checkpoint kinase 2 |
| chr5_-_19559244 | 8.17 |
ENSRNOT00000014289
ENSRNOT00000089666 |
Nsmaf
|
neutral sphingomyelinase activation associated factor |
| chr7_-_143863186 | 7.96 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr17_-_78812111 | 7.73 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr8_+_5893249 | 7.70 |
ENSRNOT00000014041
|
Mmp7
|
matrix metallopeptidase 7 |
| chr1_-_101514547 | 7.57 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr17_-_84488480 | 7.49 |
ENSRNOT00000000158
ENSRNOT00000075983 |
Nebl
|
nebulette |
| chr1_+_197999037 | 7.45 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr2_-_192671059 | 7.32 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr2_-_197860699 | 7.20 |
ENSRNOT00000028740
|
Ecm1
|
extracellular matrix protein 1 |
| chr14_+_34727915 | 7.19 |
ENSRNOT00000085089
|
Kdr
|
kinase insert domain receptor |
| chr8_-_82492243 | 7.04 |
ENSRNOT00000013852
|
Tmod3
|
tropomodulin 3 |
| chr2_+_198040536 | 6.97 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr7_+_54213319 | 6.94 |
ENSRNOT00000005286
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chrX_+_15679254 | 6.76 |
ENSRNOT00000045256
|
Plp2
|
proteolipid protein 2 |
| chr5_+_164796185 | 6.76 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr14_+_2050483 | 6.43 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chr20_-_30748784 | 6.39 |
ENSRNOT00000084391
|
Sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr19_+_37576075 | 6.38 |
ENSRNOT00000089022
|
Fam65a
|
family with sequence similarity 65, member A |
| chr8_+_80965255 | 6.29 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr6_+_93461713 | 6.13 |
ENSRNOT00000031595
|
Arid4a
|
AT-rich interaction domain 4A |
| chr1_-_88193346 | 5.99 |
ENSRNOT00000078600
|
LOC103690068
|
immortalization up-regulated protein-like |
| chr1_-_42971208 | 5.93 |
ENSRNOT00000088535
|
Gm5414
|
predicted gene 5414 |
| chr2_-_187863503 | 5.86 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr13_+_88557860 | 5.85 |
ENSRNOT00000058547
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr2_-_186480278 | 5.82 |
ENSRNOT00000031743
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chr8_+_48571323 | 5.75 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr2_+_2605996 | 5.69 |
ENSRNOT00000016372
|
Glrx
|
glutaredoxin |
| chr7_+_13062196 | 5.66 |
ENSRNOT00000000193
|
Plpp2
|
phospholipid phosphatase 2 |
| chr15_+_42653148 | 5.66 |
ENSRNOT00000022095
|
Clu
|
clusterin |
| chr9_-_119332967 | 5.58 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr2_-_29121104 | 5.20 |
ENSRNOT00000020543
|
Tnpo1
|
transportin 1 |
| chr4_-_157704596 | 5.18 |
ENSRNOT00000083016
|
Ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr1_-_228755866 | 5.17 |
ENSRNOT00000083283
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr3_-_121836086 | 5.15 |
ENSRNOT00000006113
|
Il1a
|
interleukin 1 alpha |
| chr2_-_197814808 | 5.14 |
ENSRNOT00000074156
|
Adamtsl4
|
ADAMTS-like 4 |
| chr14_+_17210733 | 5.09 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr4_-_163649637 | 5.04 |
ENSRNOT00000080873
|
LOC690020
|
similar to killer cell lectin-like receptor, subfamily A, member 17 |
| chr12_-_2568382 | 5.03 |
ENSRNOT00000035142
|
Lrrc8e
|
leucine rich repeat containing 8 family, member E |
| chr3_+_114900343 | 5.02 |
ENSRNOT00000068129
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr5_+_134691881 | 4.98 |
ENSRNOT00000091543
ENSRNOT00000067655 |
Mknk1
|
MAP kinase-interacting serine/threonine kinase 1 |
| chr2_+_227455722 | 4.97 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr1_+_197999336 | 4.92 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr9_+_95285592 | 4.83 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr3_+_110442637 | 4.83 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr1_+_91152635 | 4.82 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr2_-_158156150 | 4.82 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_-_158156444 | 4.71 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_-_63687978 | 4.64 |
ENSRNOT00000009260
|
Tbk1
|
TANK-binding kinase 1 |
| chr19_+_41631715 | 4.61 |
ENSRNOT00000022749
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr1_-_124803363 | 4.46 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr2_-_187863349 | 4.46 |
ENSRNOT00000084455
|
Lmna
|
lamin A/C |
| chr9_+_16302578 | 4.44 |
ENSRNOT00000021762
|
Gltscr1l
|
GLTSCR1-like |
| chr1_-_165997751 | 4.39 |
ENSRNOT00000050227
|
P2ry6
|
pyrimidinergic receptor P2Y6 |
| chr5_+_161889342 | 4.36 |
ENSRNOT00000040481
|
LOC100362684
|
ribosomal protein S20-like |
| chr1_+_73837944 | 4.29 |
ENSRNOT00000036413
|
Lair1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr13_-_100928811 | 4.19 |
ENSRNOT00000045326
|
Capn2
|
calpain 2 |
| chr7_-_14217778 | 4.15 |
ENSRNOT00000038961
|
Ephx3
|
epoxide hydrolase 3 |
| chr16_+_50049828 | 4.12 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr6_-_71199110 | 4.12 |
ENSRNOT00000081883
|
Prkd1
|
protein kinase D1 |
| chr18_-_28425944 | 4.08 |
ENSRNOT00000084372
|
Slc23a1
|
solute carrier family 23 member 1 |
| chr11_+_73015387 | 4.06 |
ENSRNOT00000090978
|
Gp5
|
glycoprotein V (platelet) |
| chr9_+_81402124 | 3.92 |
ENSRNOT00000046019
|
Rufy4
|
RUN and FYVE domain containing 4 |
| chr17_-_42276797 | 3.91 |
ENSRNOT00000083996
|
LOC100911664
|
uncharacterized LOC100911664 |
| chr3_-_3700200 | 3.75 |
ENSRNOT00000036231
|
AC129824.1
|
|
| chr1_-_142197182 | 3.68 |
ENSRNOT00000015521
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr1_+_185356975 | 3.56 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr20_-_10257044 | 3.49 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr4_+_44573264 | 3.49 |
ENSRNOT00000080271
|
Cav2
|
caveolin 2 |
| chr10_+_82032656 | 3.47 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr3_+_22964230 | 3.46 |
ENSRNOT00000041813
|
LOC100362149
|
ribosomal protein S20-like |
| chr20_-_3419831 | 3.45 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr1_-_214263581 | 3.38 |
ENSRNOT00000044238
|
Cdhr5
|
cadherin-related family member 5 |
| chr13_+_51384389 | 3.34 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr10_-_56491715 | 3.34 |
ENSRNOT00000020970
|
Kctd11
|
potassium channel tetramerization domain containing 11 |
| chr4_-_155690869 | 3.32 |
ENSRNOT00000012216
|
C3ar1
|
complement C3a receptor 1 |
| chr5_+_135687538 | 3.20 |
ENSRNOT00000091664
|
AC126292.2
|
|
| chr1_-_65681440 | 3.16 |
ENSRNOT00000026305
|
Zfp128
|
zinc finger protein 128 |
| chr16_+_90325304 | 3.14 |
ENSRNOT00000057310
|
Slc10a2
|
solute carrier family 10 member 2 |
| chr1_-_80864846 | 3.13 |
ENSRNOT00000074412
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr10_+_35537977 | 3.13 |
ENSRNOT00000071917
|
Rnf130
|
ring finger protein 130 |
| chr2_-_193136520 | 3.11 |
ENSRNOT00000042142
|
Kprp
|
keratinocyte proline-rich protein |
| chr1_+_72636959 | 3.07 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr3_-_81911197 | 3.04 |
ENSRNOT00000066526
|
Prdm11
|
PR/SET domain 11 |
| chr15_-_61564695 | 2.98 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr18_-_29562153 | 2.97 |
ENSRNOT00000023977
|
Cd14
|
CD14 molecule |
| chr18_+_56364620 | 2.97 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr11_+_74014983 | 2.94 |
ENSRNOT00000040884
|
NEWGENE_2724
|
glycoprotein V platelet |
| chr9_+_94425252 | 2.92 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Jun
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 21.5 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 10.2 | 40.7 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 9.5 | 28.4 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 8.5 | 25.4 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 8.3 | 25.0 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 7.0 | 21.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 7.0 | 20.9 | GO:0060101 | negative regulation of phagocytosis, engulfment(GO:0060101) |
| 6.8 | 20.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 5.8 | 17.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 4.8 | 14.3 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 4.7 | 18.8 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 4.5 | 13.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 4.5 | 13.5 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 4.5 | 17.8 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 4.3 | 12.8 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 4.1 | 12.3 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 3.9 | 11.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) regulation of DNA replication origin binding(GO:1902595) |
| 3.6 | 25.0 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 3.5 | 14.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 3.4 | 13.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 3.3 | 3.3 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 3.3 | 19.8 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 3.3 | 9.9 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 3.2 | 9.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 3.1 | 12.6 | GO:1903898 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 3.0 | 12.0 | GO:1905230 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 2.9 | 11.6 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 2.9 | 14.4 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 2.8 | 13.8 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 2.8 | 27.6 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 2.7 | 8.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 2.6 | 7.7 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 2.5 | 12.6 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 2.5 | 7.6 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 2.5 | 7.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 2.3 | 9.2 | GO:0032919 | spermine acetylation(GO:0032919) |
| 2.2 | 56.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 2.2 | 41.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 2.0 | 8.2 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 2.0 | 18.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 2.0 | 10.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 2.0 | 21.9 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 1.9 | 5.7 | GO:1902988 | regulation of neuronal signal transduction(GO:1902847) regulation of tau-protein kinase activity(GO:1902947) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.9 | 11.3 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 1.8 | 12.4 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 1.8 | 28.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 1.7 | 3.5 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 1.7 | 5.1 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 1.7 | 11.7 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.5 | 4.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 1.5 | 20.9 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 1.5 | 3.0 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 1.5 | 20.7 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 1.4 | 8.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 1.4 | 36.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 1.4 | 4.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.2 | 6.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 1.2 | 25.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 1.2 | 73.3 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 1.2 | 9.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 20.9 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 1.2 | 4.6 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 1.1 | 25.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 1.1 | 8.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 1.0 | 4.2 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 1.0 | 2.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 1.0 | 7.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 1.0 | 6.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 1.0 | 17.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.9 | 2.8 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.9 | 8.9 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.9 | 7.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.9 | 3.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.9 | 15.5 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.9 | 4.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.8 | 9.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.8 | 49.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.7 | 3.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.7 | 5.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.7 | 3.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.7 | 7.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.7 | 10.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.6 | 2.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.6 | 9.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 4.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.6 | 1.8 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.6 | 37.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.6 | 2.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.6 | 2.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.6 | 22.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.5 | 4.8 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 6.9 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.5 | 18.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.5 | 8.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.5 | 31.0 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.4 | 18.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.4 | 12.7 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.4 | 6.4 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.4 | 5.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.4 | 5.9 | GO:0045953 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of cell killing(GO:0031342) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.4 | 2.7 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.4 | 3.0 | GO:0071724 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.4 | 7.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.4 | 2.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.4 | 7.2 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.4 | 2.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.4 | 2.8 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.3 | 6.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 2.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.3 | 1.7 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.3 | 1.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) regulation of determination of dorsal identity(GO:2000015) |
| 0.3 | 2.0 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.3 | 13.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 4.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 24.6 | GO:0031016 | pancreas development(GO:0031016) |
| 0.3 | 3.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.3 | 2.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 4.9 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.3 | 2.7 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.3 | 1.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 4.9 | GO:0007567 | parturition(GO:0007567) |
| 0.3 | 0.8 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.2 | 1.9 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.2 | 2.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 1.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.2 | 2.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.2 | 1.3 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.9 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.2 | 19.1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.2 | 0.6 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 1.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 5.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 2.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 5.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 5.0 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 2.9 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 8.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 21.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.3 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 1.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 2.6 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 12.6 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.1 | 1.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.8 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.4 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 3.1 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 2.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 1.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 8.2 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.1 | 10.2 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 2.3 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 6.4 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.1 | 1.5 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 1.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.3 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 5.0 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 3.1 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 8.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.0 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 1.3 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.0 | 2.6 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.4 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 31.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 6.0 | 24.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 4.7 | 14.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 3.9 | 11.7 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 3.9 | 11.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 3.8 | 11.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 3.5 | 28.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 2.9 | 14.3 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 2.8 | 25.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 2.5 | 19.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 2.3 | 22.8 | GO:0008091 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) cuticular plate(GO:0032437) |
| 2.0 | 9.9 | GO:0097413 | Lewy body(GO:0097413) |
| 1.8 | 64.6 | GO:0008305 | integrin complex(GO:0008305) |
| 1.7 | 41.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 1.7 | 18.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.5 | 13.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 1.4 | 8.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.3 | 10.1 | GO:0005638 | lamin filament(GO:0005638) |
| 1.2 | 10.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.1 | 3.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.0 | 27.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.9 | 25.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.9 | 7.7 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.8 | 13.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.8 | 43.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.7 | 30.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.7 | 3.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.6 | 5.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.6 | 5.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.6 | 3.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.6 | 7.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.5 | 7.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 5.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 39.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.4 | 26.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.4 | 32.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.4 | 43.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.4 | 39.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.4 | 17.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 5.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.4 | 91.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 1.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 17.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 7.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 2.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 2.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 3.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 5.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 4.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 26.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 3.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 3.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 11.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 55.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.2 | 1.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 6.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 47.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 4.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 2.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 2.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 12.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 6.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.6 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.1 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 5.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 9.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 9.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 5.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 13.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 18.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 5.0 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 6.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 4.5 | 35.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 4.2 | 16.9 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 3.6 | 25.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 3.5 | 21.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 3.5 | 17.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 3.3 | 10.0 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 3.2 | 12.6 | GO:0034618 | arginine binding(GO:0034618) |
| 3.1 | 9.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 2.9 | 20.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 2.7 | 8.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 2.7 | 16.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 2.5 | 12.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 2.4 | 12.0 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 1.7 | 13.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 1.7 | 8.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.6 | 11.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 1.5 | 34.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.5 | 20.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 1.5 | 20.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.5 | 8.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.5 | 4.4 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 1.4 | 7.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.4 | 17.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 1.4 | 14.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 1.4 | 4.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.3 | 30.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 1.3 | 9.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.3 | 10.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 1.2 | 8.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 1.2 | 11.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 1.2 | 21.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.1 | 7.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 1.0 | 5.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 27.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 1.0 | 20.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.9 | 18.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.9 | 21.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.8 | 5.7 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.8 | 4.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.7 | 2.2 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.7 | 2.8 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.7 | 4.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.7 | 15.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.7 | 19.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 8.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.6 | 1.9 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.6 | 3.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 9.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.6 | 2.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.5 | 5.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.5 | 24.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.5 | 7.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 11.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.5 | 10.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.5 | 5.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.5 | 1.4 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.4 | 30.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 6.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 2.6 | GO:0032405 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.4 | 5.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 13.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.4 | 2.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 3.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.4 | 3.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.4 | 20.7 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.4 | 14.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.4 | 13.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.4 | 5.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 12.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 1.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 6.8 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.3 | 6.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 2.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 3.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.3 | 2.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 1.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.3 | 1.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 7.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.3 | 3.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 3.5 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.3 | 1.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 4.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 8.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 34.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 32.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 11.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 2.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 10.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 3.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 2.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 4.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 3.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 4.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 5.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 46.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 1.9 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.1 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 60.8 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.1 | 2.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 6.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 17.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 21.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 7.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 14.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 5.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 4.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 3.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 4.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 4.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.7 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 9.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 73.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.9 | 27.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.9 | 32.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.3 | 48.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 1.2 | 28.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 1.1 | 30.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 1.1 | 20.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 1.0 | 21.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.9 | 10.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.9 | 11.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.9 | 33.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.8 | 25.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.7 | 13.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.6 | 35.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.6 | 50.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.6 | 5.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.5 | 15.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.5 | 9.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.5 | 6.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.5 | 8.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.5 | 19.4 | PID ATM PATHWAY | ATM pathway |
| 0.4 | 4.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.4 | 8.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.4 | 12.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 8.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 13.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 12.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 2.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 3.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 4.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 3.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 7.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 9.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 5.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 5.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 5.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 9.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 15.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 25.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 1.6 | 12.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 1.4 | 9.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.4 | 45.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 1.3 | 47.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 1.1 | 14.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.1 | 14.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.0 | 27.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.0 | 86.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.9 | 11.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.8 | 5.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.8 | 7.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.8 | 31.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.7 | 8.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.7 | 25.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.7 | 28.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.7 | 11.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.6 | 21.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 11.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.6 | 7.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 1.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.6 | 17.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.5 | 3.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.5 | 9.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 4.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 4.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 23.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.4 | 13.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 4.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 15.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.3 | 16.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 12.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.3 | 5.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 5.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 7.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 5.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.3 | 4.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 25.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 8.9 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.2 | 5.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 9.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 9.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.2 | 2.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 1.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 21.0 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 9.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 10.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 5.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 2.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |