Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
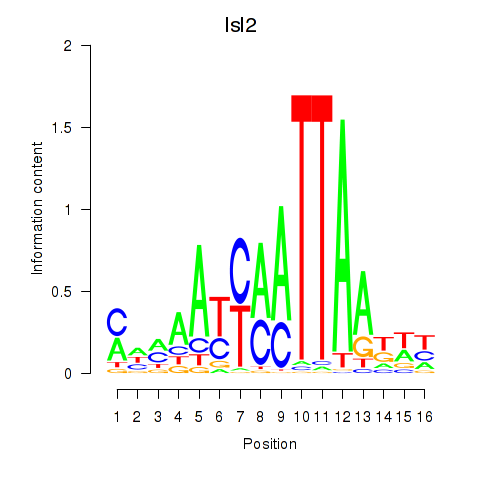
Results for Isl2
Z-value: 0.56
Transcription factors associated with Isl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl2
|
ENSRNOG00000015336 | ISL LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl2 | rn6_v1_chr8_+_60117729_60117729 | 0.34 | 2.8e-10 | Click! |
Activity profile of Isl2 motif
Sorted Z-values of Isl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_28164326 | 9.78 |
ENSRNOT00000088165
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr5_+_28485619 | 9.60 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr16_-_49574314 | 9.49 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr3_+_143119687 | 9.25 |
ENSRNOT00000006608
|
Cst12
|
cystatin 12 |
| chr5_-_115387377 | 9.12 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr10_+_84135116 | 8.61 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr1_+_128637049 | 8.37 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr1_-_227457629 | 7.71 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr16_-_24951612 | 7.63 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr4_+_29535852 | 7.35 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr14_+_34389991 | 7.06 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr1_+_128606770 | 6.83 |
ENSRNOT00000073355
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_-_42002819 | 6.60 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr4_-_163762434 | 6.49 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr1_-_237910012 | 6.37 |
ENSRNOT00000023664
|
Anxa1
|
annexin A1 |
| chr14_-_21748356 | 6.36 |
ENSRNOT00000002670
|
Cabs1
|
calcium binding protein, spermatid associated 1 |
| chr20_+_17750744 | 6.24 |
ENSRNOT00000000745
|
RGD1559903
|
similar to RIKEN cDNA 1700049L16 |
| chr9_-_105376935 | 6.24 |
ENSRNOT00000043386
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr13_-_74005486 | 6.21 |
ENSRNOT00000090173
|
AABR07021465.1
|
|
| chr14_+_17228856 | 5.99 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr13_-_105684374 | 5.99 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr13_-_83457888 | 5.93 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr1_+_101603222 | 5.79 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr4_-_164015365 | 5.68 |
ENSRNOT00000078121
|
Ly49si2
|
immunoreceptor Ly49si2 |
| chr4_+_1470716 | 5.62 |
ENSRNOT00000044223
|
Olr1235
|
olfactory receptor 1235 |
| chr19_-_42180362 | 5.60 |
ENSRNOT00000089515
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr5_+_133896141 | 5.60 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr9_-_104870225 | 5.54 |
ENSRNOT00000065171
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr1_-_54854353 | 5.49 |
ENSRNOT00000072895
|
Smok2a
|
sperm motility kinase 2A |
| chr9_-_104870382 | 5.46 |
ENSRNOT00000084398
|
Slco6d1
|
solute carrier organic anion transporter family, member 6d1 |
| chr17_+_47302272 | 5.22 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chr16_+_72010106 | 5.15 |
ENSRNOT00000058330
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr5_-_113880911 | 5.13 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr17_+_89452814 | 5.11 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr15_+_80040842 | 5.08 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr1_-_14117021 | 5.03 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr8_+_79638696 | 5.02 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr17_-_42031265 | 4.78 |
ENSRNOT00000068021
|
Dcdc2
|
doublecortin domain containing 2 |
| chr3_-_37854561 | 4.68 |
ENSRNOT00000076095
|
Neb
|
nebulin |
| chr1_+_201337416 | 4.67 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr9_-_80167033 | 4.57 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr14_-_77810147 | 4.56 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr2_+_2729829 | 4.55 |
ENSRNOT00000017068
|
Spata9
|
spermatogenesis associated 9 |
| chr5_-_129175597 | 4.51 |
ENSRNOT00000012112
|
LOC689589
|
hypothetical protein LOC689589 |
| chr8_+_117170620 | 4.50 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr6_+_101288951 | 4.48 |
ENSRNOT00000046901
|
RGD1562540
|
RGD1562540 |
| chr17_+_47301511 | 4.34 |
ENSRNOT00000092051
ENSRNOT00000087178 |
Nme8
|
NME/NM23 family member 8 |
| chr9_-_104467973 | 4.33 |
ENSRNOT00000026099
|
Slco6b1
|
solute carrier organic anion transporter family member 6B1 |
| chrX_-_159841072 | 4.29 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr7_-_124929025 | 4.21 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr9_+_53013413 | 4.13 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chrX_+_25737292 | 4.05 |
ENSRNOT00000004893
|
RGD1563947
|
similar to RIKEN cDNA 4933400A11 |
| chr1_-_67284864 | 4.04 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr9_-_80166807 | 4.02 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr8_+_22559098 | 4.01 |
ENSRNOT00000041091
|
LOC691141
|
hypothetical protein LOC691141 |
| chr17_-_14627937 | 3.98 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr15_-_35153804 | 3.86 |
ENSRNOT00000050660
|
Gzmn
|
granzyme N |
| chr2_-_157035483 | 3.80 |
ENSRNOT00000076491
|
LOC304239
|
similar to RalA binding protein 1 |
| chrX_-_63999622 | 3.78 |
ENSRNOT00000090902
|
LOC103694487
|
protein gar2-like |
| chr10_-_13716931 | 3.73 |
ENSRNOT00000060818
|
Abca17
|
ATP-binding cassette, subfamily A (ABC1), member 17 |
| chr1_+_253221812 | 3.67 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chrX_-_124162058 | 3.65 |
ENSRNOT00000071436
|
LOC367975
|
similar to B-cell translocation gene 1 |
| chr5_+_103251986 | 3.61 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr10_-_10767389 | 3.58 |
ENSRNOT00000066754
|
Smim22
|
small integral membrane protein 22 |
| chr1_+_18491384 | 3.49 |
ENSRNOT00000079138
ENSRNOT00000014917 |
Lama2
|
laminin subunit alpha 2 |
| chr4_-_176606382 | 3.49 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr1_+_252284464 | 3.47 |
ENSRNOT00000027969
|
Lipf
|
lipase F, gastric type |
| chr6_-_99273033 | 3.40 |
ENSRNOT00000088808
|
Tex21
|
testis expressed 21 |
| chr19_-_49510901 | 3.39 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr9_+_8054466 | 3.33 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr2_-_181900856 | 3.32 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chrX_+_84064427 | 3.29 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr10_+_56524468 | 3.26 |
ENSRNOT00000022041
|
Gps2
|
G protein pathway suppressor 2 |
| chrX_-_98095663 | 3.19 |
ENSRNOT00000004491
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr16_-_29936307 | 3.15 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr4_+_61814974 | 3.09 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr7_-_143167772 | 3.06 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chrX_+_77143892 | 3.04 |
ENSRNOT00000085227
|
Atp7a
|
ATPase copper transporting alpha |
| chr2_-_216348194 | 3.02 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr6_-_94932806 | 2.98 |
ENSRNOT00000006346
|
Ccdc175
|
coiled-coil domain containing 175 |
| chr7_+_38858062 | 2.91 |
ENSRNOT00000006234
|
Kera
|
keratocan |
| chr10_+_61685645 | 2.89 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr9_+_60900213 | 2.88 |
ENSRNOT00000017859
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr9_+_8052210 | 2.86 |
ENSRNOT00000073659
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr9_-_27452902 | 2.85 |
ENSRNOT00000018325
|
Gsta1
|
glutathione S-transferase alpha 1 |
| chr3_+_8430829 | 2.85 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr1_+_79631668 | 2.82 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr1_-_127292090 | 2.81 |
ENSRNOT00000016959
|
Lrrk1
|
leucine-rich repeat kinase 1 |
| chr14_-_21299068 | 2.79 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr2_+_122877286 | 2.76 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr9_-_70449796 | 2.69 |
ENSRNOT00000086391
|
Mdh1b
|
malate dehydrogenase 1B |
| chr11_-_28900376 | 2.69 |
ENSRNOT00000061606
|
Krtap16-5
|
keratin associated protein 16-5 |
| chr14_-_21810314 | 2.68 |
ENSRNOT00000002671
|
Csn3
|
casein kappa |
| chr10_-_98469799 | 2.65 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr5_-_108747666 | 2.63 |
ENSRNOT00000072732
|
Zfp353
|
zinc finger protein 353 |
| chr10_-_29026002 | 2.62 |
ENSRNOT00000005070
|
Pttg1
|
pituitary tumor-transforming 1 |
| chr11_+_15081774 | 2.58 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr4_-_184096806 | 2.52 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr4_-_164899041 | 2.51 |
ENSRNOT00000090316
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr8_-_40883880 | 2.50 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr2_-_192288568 | 2.49 |
ENSRNOT00000067346
|
AABR07012291.2
|
|
| chr7_+_2300434 | 2.47 |
ENSRNOT00000071785
|
AABR07055413.1
|
|
| chr2_-_216382244 | 2.41 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr4_-_50693869 | 2.39 |
ENSRNOT00000064228
|
Rnf148
|
ring finger protein 148 |
| chr4_-_164304532 | 2.38 |
ENSRNOT00000089484
|
Ly49i5
|
Ly49 inhibitory receptor 5 |
| chr1_+_80028928 | 2.38 |
ENSRNOT00000012082
|
Fbxo46
|
F-box protein 46 |
| chr16_+_39145230 | 2.38 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr5_+_142986526 | 2.34 |
ENSRNOT00000012811
|
Rspo1
|
R-spondin 1 |
| chr10_+_87759769 | 2.32 |
ENSRNOT00000017378
ENSRNOT00000046526 |
Krtap9-1
|
keratin associated protein 9-1 |
| chr1_+_225163391 | 2.31 |
ENSRNOT00000027305
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr8_-_17524839 | 2.30 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr10_-_82117109 | 2.29 |
ENSRNOT00000079711
|
Abcc3
|
ATP binding cassette subfamily C member 3 |
| chr3_+_48626038 | 2.29 |
ENSRNOT00000009697
|
Gca
|
grancalcin |
| chr13_-_82295123 | 2.25 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr3_+_93909156 | 2.24 |
ENSRNOT00000090365
|
Lmo2
|
LIM domain only 2 |
| chr6_-_140880070 | 2.20 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr12_-_30491102 | 2.20 |
ENSRNOT00000001224
|
Sumf2
|
sulfatase modifying factor 2 |
| chrX_+_23414354 | 2.18 |
ENSRNOT00000031235
|
Cldn34a
|
claudin 34A |
| chr6_+_76079880 | 2.14 |
ENSRNOT00000009304
|
RGD1305089
|
similar to 1110008L16Rik protein |
| chr4_+_22898527 | 2.09 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr2_+_30664639 | 2.08 |
ENSRNOT00000076372
ENSRNOT00000076294 ENSRNOT00000076434 ENSRNOT00000076484 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr2_-_27343998 | 2.08 |
ENSRNOT00000091418
|
Polk
|
DNA polymerase kappa |
| chr1_+_61382118 | 2.05 |
ENSRNOT00000050119
|
Zfp52
|
zinc finger protein 52 |
| chr3_-_57607683 | 2.03 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr14_-_21709084 | 2.01 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr5_+_144106802 | 2.00 |
ENSRNOT00000035637
ENSRNOT00000079779 |
Lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr14_+_23405717 | 2.00 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr4_-_155690869 | 1.98 |
ENSRNOT00000012216
|
C3ar1
|
complement C3a receptor 1 |
| chr1_-_78180216 | 1.97 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr4_-_80395502 | 1.94 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr15_-_32925673 | 1.93 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chr10_-_34242985 | 1.88 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr8_+_104106740 | 1.87 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr3_-_80933283 | 1.86 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr12_+_41316764 | 1.85 |
ENSRNOT00000090867
|
Oas3
|
2'-5'-oligoadenylate synthetase 3 |
| chr7_+_16404755 | 1.83 |
ENSRNOT00000044977
|
LOC108349290
|
olfactory receptor 6C70-like |
| chr16_+_29674793 | 1.80 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr7_-_139649286 | 1.78 |
ENSRNOT00000080056
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr3_+_102456938 | 1.75 |
ENSRNOT00000051827
|
Olr748
|
olfactory receptor 748 |
| chr1_-_227965386 | 1.74 |
ENSRNOT00000028491
|
Ms4a2
|
membrane spanning 4-domains A2 |
| chr15_-_36798814 | 1.73 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr14_-_21758788 | 1.68 |
ENSRNOT00000038520
|
2310003L06Rik
|
RIKEN cDNA 2310003L06 gene |
| chr5_-_162751128 | 1.68 |
ENSRNOT00000068281
|
RGD1559644
|
similar to novel protein similar to esterases |
| chrX_-_64715823 | 1.66 |
ENSRNOT00000076297
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr4_-_66955732 | 1.66 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr8_+_127702534 | 1.66 |
ENSRNOT00000075793
|
Ctdspl
|
CTD small phosphatase like |
| chr11_+_81358592 | 1.66 |
ENSRNOT00000002487
|
Rfc4
|
replication factor C subunit 4 |
| chr3_-_431933 | 1.63 |
ENSRNOT00000033653
|
Spopl
|
speckle type BTB/POZ protein like |
| chr8_-_84835060 | 1.63 |
ENSRNOT00000007867
|
Lrrc1
|
leucine rich repeat containing 1 |
| chrX_+_124631881 | 1.63 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr4_-_162649618 | 1.62 |
ENSRNOT00000050861
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr2_-_192481273 | 1.62 |
ENSRNOT00000012135
|
Sprr2d
|
small proline-rich protein 2D |
| chr1_-_90057923 | 1.55 |
ENSRNOT00000028674
|
Pdcd2l
|
programmed cell death 2-like |
| chr1_-_126211439 | 1.54 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr2_+_30664217 | 1.53 |
ENSRNOT00000076786
|
Ak6
|
adenylate kinase 6 |
| chr15_+_39779648 | 1.51 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr4_+_163349125 | 1.50 |
ENSRNOT00000084823
|
Klre1
|
killer cell lectin-like receptor, family E, member 1 |
| chr17_-_10001901 | 1.43 |
ENSRNOT00000082836
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr3_-_147263275 | 1.42 |
ENSRNOT00000029620
|
Psmf1
|
proteasome inhibitor subunit 1 |
| chr8_-_128026841 | 1.41 |
ENSRNOT00000018341
|
Myd88
|
myeloid differentiation primary response 88 |
| chr8_+_40707068 | 1.38 |
ENSRNOT00000074529
|
Olr1214
|
olfactory receptor 1214 |
| chr4_+_156752082 | 1.38 |
ENSRNOT00000084588
ENSRNOT00000068407 |
Cd163
|
CD163 molecule |
| chr3_-_102484517 | 1.37 |
ENSRNOT00000006522
|
Olr750
|
olfactory receptor 750 |
| chr3_-_165700489 | 1.35 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr17_+_84881414 | 1.33 |
ENSRNOT00000034157
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr17_+_45467015 | 1.32 |
ENSRNOT00000081408
|
Gpx5
|
glutathione peroxidase 5 |
| chr1_-_755645 | 1.31 |
ENSRNOT00000073546
|
Vom2r6
|
vomeronasal 2 receptor, 6 |
| chr7_-_143497108 | 1.29 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr3_+_160092975 | 1.28 |
ENSRNOT00000080707
|
Pkig
|
cAMP-dependent protein kinase inhibitor gamma |
| chr2_-_198706428 | 1.28 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr8_+_61671513 | 1.28 |
ENSRNOT00000091128
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr4_+_149908375 | 1.27 |
ENSRNOT00000019504
|
LOC100909657
|
uncharacterized LOC100909657 |
| chr4_-_170186942 | 1.25 |
ENSRNOT00000082235
|
Wbp11l1
|
WW domain-binding protein 11-like 1 |
| chr2_+_88217188 | 1.25 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chr20_-_13994794 | 1.24 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr1_-_167698263 | 1.24 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr4_+_70614524 | 1.22 |
ENSRNOT00000041100
|
Prss3
|
protease, serine 3 |
| chr10_+_95770154 | 1.21 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr1_+_169115981 | 1.21 |
ENSRNOT00000067478
|
Olr135
|
olfactory receptor 135 |
| chr3_+_63379031 | 1.19 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr4_-_59014416 | 1.18 |
ENSRNOT00000049811
|
RGD1561636
|
similar to 60S ribosomal protein L38 |
| chr1_+_227892956 | 1.17 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr15_+_108318664 | 1.17 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr14_-_24123253 | 1.13 |
ENSRNOT00000002743
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chr15_-_36451720 | 1.13 |
ENSRNOT00000075752
|
LOC691695
|
similar to granzyme C |
| chr6_-_105214329 | 1.09 |
ENSRNOT00000008954
|
Cox16
|
COX16, cytochrome c oxidase assembly homolog |
| chr8_-_53277859 | 1.09 |
ENSRNOT00000009574
|
Htr3b
|
5-hydroxytryptamine receptor 3B |
| chr9_-_14550625 | 1.06 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_+_18068060 | 1.05 |
ENSRNOT00000065474
|
Vom1r107
|
vomeronasal 1 receptor 107 |
| chr1_-_238376841 | 1.04 |
ENSRNOT00000076393
|
Tmc1
|
transmembrane channel-like 1 |
| chr15_-_35394792 | 1.04 |
ENSRNOT00000028058
|
Gzmc
|
granzyme C |
| chr19_-_56799445 | 1.04 |
ENSRNOT00000024491
|
RGD1563991
|
similar to TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr4_-_148437961 | 1.02 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr7_+_122203532 | 1.02 |
ENSRNOT00000025382
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr4_-_164536556 | 1.01 |
ENSRNOT00000087796
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr10_+_106812739 | 1.01 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr3_+_112346627 | 1.00 |
ENSRNOT00000074392
|
Snap23
|
synaptosomal-associated protein 23 |
| chr5_+_124476168 | 0.99 |
ENSRNOT00000077754
|
RGD1564074
|
similar to novel protein |
| chr5_+_163867961 | 0.97 |
ENSRNOT00000081495
|
AABR07050399.1
|
|
| chr9_+_95161157 | 0.96 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 1.3 | 6.4 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 1.2 | 3.7 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.0 | 3.1 | GO:0016107 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 1.0 | 5.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.9 | 2.8 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.9 | 7.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.9 | 9.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.8 | 3.0 | GO:0051542 | negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.7 | 2.8 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.7 | 2.0 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.7 | 2.6 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.6 | 9.8 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.5 | 2.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.5 | 10.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.5 | 6.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.5 | 1.5 | GO:0002856 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.5 | 5.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.5 | 1.4 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.5 | 9.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.4 | 1.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.4 | 3.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 2.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.4 | 4.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.4 | 1.4 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.3 | 1.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 2.9 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 8.6 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 3.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.3 | 6.0 | GO:0010818 | T cell chemotaxis(GO:0010818) induction of positive chemotaxis(GO:0050930) positive regulation of myoblast fusion(GO:1901741) |
| 0.3 | 1.9 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 2.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 0.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 2.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 | 2.3 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.3 | 0.5 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.3 | 1.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.3 | 1.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 4.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 0.7 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 | 0.7 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 1.5 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.2 | 2.8 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 1.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 1.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 1.9 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.2 | 2.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 2.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 1.9 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.2 | 2.8 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.2 | 2.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.7 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.2 | 1.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 1.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 1.2 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.9 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 1.0 | GO:0048227 | regulation of Rab protein signal transduction(GO:0032483) plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.0 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 4.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 2.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.3 | GO:0089709 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 1.0 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 4.3 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 3.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.5 | GO:0061364 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 2.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 1.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.4 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.1 | 0.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.6 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 4.0 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 2.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.6 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.2 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 10.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.3 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 2.7 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.2 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 1.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.8 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 1.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:1902961 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.8 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 1.4 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 1.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.0 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 3.2 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.5 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.5 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.0 | 1.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.6 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.9 | GO:0098869 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.7 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 4.3 | GO:0007283 | spermatogenesis(GO:0007283) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.6 | GO:0097598 | outer dynein arm(GO:0036157) sperm cytoplasmic droplet(GO:0097598) |
| 1.4 | 8.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.3 | 5.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.7 | 6.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.6 | 10.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 2.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.3 | 4.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 5.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 2.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 1.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 5.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 3.7 | GO:1990023 | mitotic spindle pole(GO:0097431) mitotic spindle midzone(GO:1990023) |
| 0.2 | 1.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 6.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 0.8 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.2 | 2.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 3.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 5.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 2.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 6.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 4.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 4.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 5.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 3.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 3.6 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.3 | 6.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.2 | 6.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 3.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 1.0 | 8.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.9 | 7.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.8 | 2.5 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.8 | 9.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.7 | 10.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.7 | 2.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.7 | 2.0 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.7 | 2.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.7 | 9.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 5.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 3.0 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 0.6 | 3.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.5 | 9.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.5 | 3.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.5 | 1.4 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.4 | 2.2 | GO:0004140 | adenylate kinase activity(GO:0004017) dephospho-CoA kinase activity(GO:0004140) |
| 0.4 | 1.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 2.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 2.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.4 | 1.8 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 1.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.3 | 0.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 2.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 2.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.3 | 3.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 2.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.3 | 2.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 2.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.2 | 2.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 1.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 9.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 4.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 11.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 0.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 0.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.5 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 2.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 7.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 2.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.4 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 7.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 17.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 4.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 9.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 5.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 4.2 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 3.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 7.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 8.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 1.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 4.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 4.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 2.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 4.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 10.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 4.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 5.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 3.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.1 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |