Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
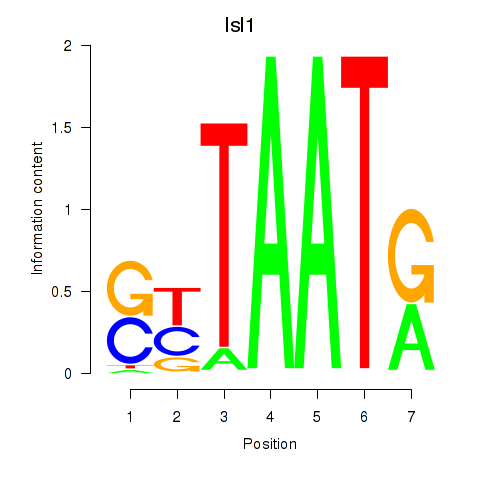
Results for Isl1
Z-value: 0.34
Transcription factors associated with Isl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl1
|
ENSRNOG00000012556 | ISL LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl1 | rn6_v1_chr2_-_48501436_48501436 | 0.04 | 5.0e-01 | Click! |
Activity profile of Isl1 motif
Sorted Z-values of Isl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_12820466 | 13.00 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr9_-_26707571 | 11.14 |
ENSRNOT00000080948
|
AABR07067023.1
|
|
| chrX_-_32153794 | 9.71 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chrX_+_23081125 | 8.91 |
ENSRNOT00000071639
|
AABR07037520.1
|
|
| chr14_-_64476796 | 7.78 |
ENSRNOT00000029104
|
Gba3
|
glucosidase, beta, acid 3 |
| chr14_-_19191863 | 6.04 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr10_-_31359699 | 6.01 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr14_+_20266891 | 5.16 |
ENSRNOT00000004174
|
Gc
|
group specific component |
| chr9_-_82336806 | 5.03 |
ENSRNOT00000024667
|
Slc23a3
|
solute carrier family 23, member 3 |
| chrX_-_32095867 | 4.82 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr8_+_57936650 | 4.67 |
ENSRNOT00000089686
|
Exph5
|
exophilin 5 |
| chr4_+_157836912 | 4.58 |
ENSRNOT00000067271
|
Scnn1a
|
sodium channel epithelial 1 alpha subunit |
| chr1_-_102826965 | 4.50 |
ENSRNOT00000078692
|
Saa4
|
serum amyloid A4 |
| chr3_+_54253949 | 4.48 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr1_-_43638161 | 4.41 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_117421604 | 4.38 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr15_-_52320385 | 4.09 |
ENSRNOT00000067776
|
Dmtn
|
dematin actin binding protein |
| chr2_-_100372252 | 3.84 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr6_-_46631983 | 3.73 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr18_-_4294136 | 3.39 |
ENSRNOT00000091909
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr9_-_71900044 | 3.27 |
ENSRNOT00000020322
|
Idh1
|
isocitrate dehydrogenase (NADP(+)) 1, cytosolic |
| chr14_-_5101177 | 3.15 |
ENSRNOT00000002888
|
Lrrc8d
|
leucine rich repeat containing 8 family, member D |
| chr2_+_83393282 | 2.69 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr11_-_27971359 | 2.39 |
ENSRNOT00000085629
ENSRNOT00000051060 ENSRNOT00000042581 ENSRNOT00000050073 ENSRNOT00000081066 |
Grik1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr7_+_28654733 | 2.30 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr18_-_28017925 | 1.97 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr7_+_83113672 | 1.78 |
ENSRNOT00000006783
|
Trhr
|
thyrotropin releasing hormone receptor |
| chr15_+_62406873 | 1.68 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr19_-_58735173 | 1.66 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr15_+_12827707 | 1.63 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr17_-_77527894 | 1.57 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr15_-_29761117 | 1.50 |
ENSRNOT00000075194
|
AABR07017658.2
|
|
| chr3_-_44177689 | 1.36 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr15_+_31417147 | 1.34 |
ENSRNOT00000092182
|
AABR07017824.2
|
|
| chr14_-_3288017 | 1.33 |
ENSRNOT00000080452
|
LOC689986
|
hypothetical protein LOC689986 |
| chr3_-_25212049 | 1.26 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr4_+_88119838 | 1.25 |
ENSRNOT00000073173
|
Vom1r82
|
vomeronasal 1 receptor 82 |
| chr15_+_28023018 | 1.21 |
ENSRNOT00000090272
|
Rnase4
|
ribonuclease A family member 4 |
| chr20_-_48503898 | 1.20 |
ENSRNOT00000073091
|
Wasf1
|
WAS protein family, member 1 |
| chr3_-_71798531 | 1.19 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr14_+_69800156 | 1.08 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr15_+_31395772 | 1.06 |
ENSRNOT00000075087
|
AABR07017824.1
|
|
| chrX_-_128268285 | 1.00 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr2_+_66940057 | 1.00 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr5_+_126698582 | 0.96 |
ENSRNOT00000012427
|
Cdcp2
|
CUB domain containing protein 2 |
| chr13_-_102942863 | 0.94 |
ENSRNOT00000003198
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr10_-_21265026 | 0.89 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr5_-_128446725 | 0.83 |
ENSRNOT00000011272
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr2_-_157759819 | 0.81 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr18_-_26656879 | 0.80 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr13_-_89242443 | 0.72 |
ENSRNOT00000029202
|
Atf6
|
activating transcription factor 6 |
| chrX_+_153677811 | 0.72 |
ENSRNOT00000072808
|
AABR07042361.1
|
|
| chr19_-_37528011 | 0.72 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr15_-_29903755 | 0.71 |
ENSRNOT00000066802
|
AABR07017669.1
|
|
| chr15_+_30750093 | 0.69 |
ENSRNOT00000071830
|
AABR07017763.1
|
|
| chr5_+_172364421 | 0.63 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr4_-_167089055 | 0.62 |
ENSRNOT00000050409
|
Tas2r113
|
taste receptor, type 2, member 113 |
| chr15_+_108318664 | 0.61 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr20_+_9948908 | 0.56 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr5_-_28130803 | 0.51 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr2_+_34312766 | 0.50 |
ENSRNOT00000060962
|
Cenpk
|
centromere protein K |
| chr10_-_64657089 | 0.49 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr5_-_28131133 | 0.48 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr10_-_87286387 | 0.48 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr20_+_48503973 | 0.45 |
ENSRNOT00000064081
|
Cdc40
|
cell division cycle 40 |
| chr4_-_7885301 | 0.45 |
ENSRNOT00000035034
|
Rint1
|
RAD50 interactor 1 |
| chr3_+_21114856 | 0.44 |
ENSRNOT00000044947
|
Olr423
|
olfactory receptor 423 |
| chr1_+_168519499 | 0.38 |
ENSRNOT00000045286
|
Olr98
|
olfactory receptor 98 |
| chr20_+_48504264 | 0.37 |
ENSRNOT00000087740
|
Cdc40
|
cell division cycle 40 |
| chr3_-_76187045 | 0.36 |
ENSRNOT00000075650
|
LOC686683
|
similar to olfactory receptor 73 |
| chr3_-_161819029 | 0.34 |
ENSRNOT00000091834
|
LOC102555457
|
engulfment and cell motility protein 2-like |
| chr6_-_117972898 | 0.34 |
ENSRNOT00000032968
|
AABR07065265.1
|
|
| chr15_-_45927804 | 0.28 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr15_+_56666012 | 0.27 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr3_-_76085971 | 0.26 |
ENSRNOT00000007679
|
Olr601
|
olfactory receptor 601 |
| chr15_-_28786094 | 0.25 |
ENSRNOT00000044250
|
Olr1639
|
olfactory receptor 1639 |
| chr4_+_57883849 | 0.17 |
ENSRNOT00000013880
|
Cpa4
|
carboxypeptidase A4 |
| chr5_+_145311375 | 0.14 |
ENSRNOT00000019224
|
Smim12
|
small integral membrane protein 12 |
| chr10_-_34333305 | 0.14 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr1_+_168575090 | 0.13 |
ENSRNOT00000048299
|
Olr103
|
olfactory receptor 103 |
| chr2_+_4989295 | 0.11 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr16_-_8685529 | 0.09 |
ENSRNOT00000092751
|
Slc18a3
|
solute carrier family 18 member A3 |
| chr13_+_56513286 | 0.09 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr1_+_85213652 | 0.08 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chrX_-_63182385 | 0.04 |
ENSRNOT00000076613
|
Zfx
|
zinc finger protein X-linked |
| chr14_+_22192970 | 0.03 |
ENSRNOT00000041514
ENSRNOT00000002704 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 1.9 | 7.8 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.6 | 4.8 | GO:0015827 | angiotensin-mediated drinking behavior(GO:0003051) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 1.5 | 4.4 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 1.1 | 9.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 1.0 | 4.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.0 | 6.0 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) response to platinum ion(GO:0070541) |
| 0.9 | 3.7 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.9 | 7.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 3.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) glyoxylate metabolic process(GO:0046487) |
| 0.5 | 4.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.5 | 2.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.4 | 1.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 4.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 5.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 2.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 2.4 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.2 | 0.6 | GO:2000974 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.2 | 1.7 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.2 | 0.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.2 | 1.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.4 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 4.6 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 1.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.0 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 4.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.6 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 2.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.6 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 3.1 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 0.1 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 3.8 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.4 | 4.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 2.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 4.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 4.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 11.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 5.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 3.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.1 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 7.8 | GO:0005768 | endosome(GO:0005768) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 2.6 | 13.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 2.6 | 7.8 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 1.5 | 4.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 1.1 | 3.3 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.9 | 4.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.9 | 5.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.5 | 4.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.4 | 1.2 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.3 | 2.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 6.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 3.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.1 | 4.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 3.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 3.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 1.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 6.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 6.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 7.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 5.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 13.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 5.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 4.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |