Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
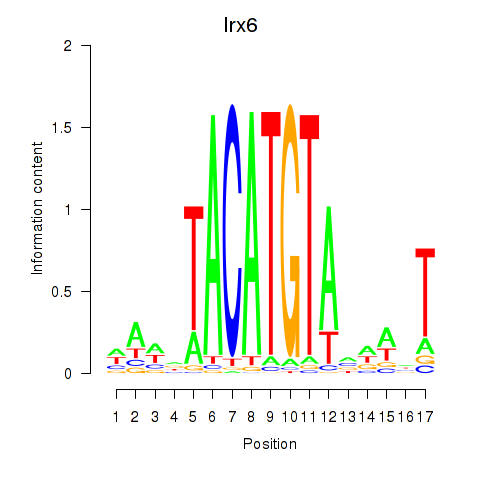
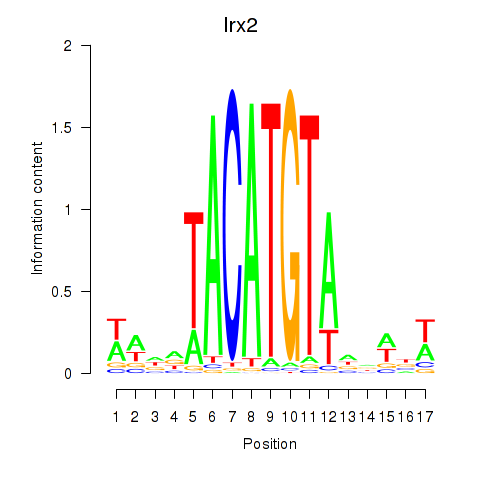
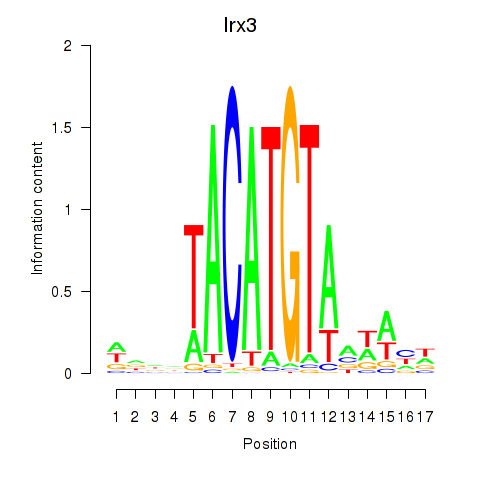
Results for Irx6_Irx2_Irx3
Z-value: 0.68
Transcription factors associated with Irx6_Irx2_Irx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx6
|
ENSRNOG00000011037 | iroquois homeobox 6 |
|
Irx2
|
ENSRNOG00000012742 | iroquois homeobox 2 |
|
Irx3
|
ENSRNOG00000011533 | iroquois homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx6 | rn6_v1_chr19_-_15733412_15733412 | -0.39 | 2.5e-13 | Click! |
| Irx3 | rn6_v1_chr19_-_15840990_15840990 | -0.26 | 3.2e-06 | Click! |
| Irx2 | rn6_v1_chr1_-_33275540_33275540 | -0.14 | 9.4e-03 | Click! |
Activity profile of Irx6_Irx2_Irx3 motif
Sorted Z-values of Irx6_Irx2_Irx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_14157141 | 25.94 |
ENSRNOT00000079034
|
Deup1
|
deuterosome assembly protein 1 |
| chr1_+_83103925 | 21.88 |
ENSRNOT00000047540
ENSRNOT00000028196 |
Cyp2b2
|
cytochrome P450, family 2, subfamily b, polypeptide 2 |
| chr1_+_53531076 | 14.77 |
ENSRNOT00000018015
|
Tcp10b
|
t-complex protein 10b |
| chr13_+_89597138 | 14.60 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr1_-_258766881 | 14.50 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr4_+_14109864 | 13.83 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr2_+_193724248 | 13.44 |
ENSRNOT00000025249
|
Rptn
|
repetin |
| chr7_-_2431197 | 12.81 |
ENSRNOT00000003498
|
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr7_+_99142450 | 11.55 |
ENSRNOT00000079036
ENSRNOT00000091923 |
LOC108348266
|
cytochrome P450 2B1 |
| chr16_-_82288022 | 10.97 |
ENSRNOT00000078609
|
Spaca7
|
sperm acrosome associated 7 |
| chr2_-_173563273 | 10.38 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr18_+_35574002 | 10.29 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr18_+_35121967 | 10.19 |
ENSRNOT00000017522
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr16_+_72086878 | 9.69 |
ENSRNOT00000023756
ENSRNOT00000078085 |
Adam3a
|
ADAM metallopeptidase domain 3A |
| chr3_-_46601409 | 9.63 |
ENSRNOT00000079261
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr13_-_56958549 | 9.42 |
ENSRNOT00000017293
ENSRNOT00000083912 |
RGD1564614
|
similar to complement factor H-related protein |
| chrX_+_105011489 | 9.14 |
ENSRNOT00000085068
|
Arl13a
|
ADP ribosylation factor like GTPase 13A |
| chr2_+_35935670 | 9.08 |
ENSRNOT00000076875
ENSRNOT00000075753 |
LOC108348103
|
serine protease inhibitor Kazal-type 5-like |
| chr9_+_24066303 | 8.96 |
ENSRNOT00000018163
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr1_-_164142206 | 8.88 |
ENSRNOT00000081669
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr9_+_37727942 | 8.87 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr10_-_107114271 | 8.87 |
ENSRNOT00000004035
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr4_+_14039977 | 8.62 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr5_-_39611053 | 8.55 |
ENSRNOT00000046595
|
Fhl5
|
four and a half LIM domains 5 |
| chr15_-_49505553 | 8.51 |
ENSRNOT00000028974
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr14_+_22375955 | 8.50 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_-_219312240 | 8.34 |
ENSRNOT00000066691
|
RGD1307603
|
similar to hypothetical protein MGC37914 |
| chr2_+_104744461 | 8.05 |
ENSRNOT00000016083
ENSRNOT00000082627 |
Cp
|
ceruloplasmin |
| chr16_+_50179458 | 8.00 |
ENSRNOT00000041946
|
F11
|
coagulation factor XI |
| chr9_+_16139101 | 7.99 |
ENSRNOT00000070803
|
LOC100910668
|
uncharacterized LOC100910668 |
| chr9_+_14951047 | 7.73 |
ENSRNOT00000074267
|
LOC100911489
|
uncharacterized LOC100911489 |
| chr7_+_40318490 | 7.38 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr3_+_1385654 | 7.21 |
ENSRNOT00000091805
ENSRNOT00000007593 |
Il36rn
|
interleukin 36 receptor antagonist |
| chr18_+_3861539 | 7.05 |
ENSRNOT00000015363
|
Lama3
|
laminin subunit alpha 3 |
| chr17_+_57033573 | 6.88 |
ENSRNOT00000020160
|
Crem
|
cAMP responsive element modulator |
| chr1_+_229267916 | 6.75 |
ENSRNOT00000073717
ENSRNOT00000082670 ENSRNOT00000076941 |
LOC100910851
|
serine protease inhibitor Kazal-type 5-like |
| chr8_-_111761871 | 6.74 |
ENSRNOT00000056483
|
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr17_-_69711689 | 6.57 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr3_-_134462653 | 6.37 |
ENSRNOT00000033562
|
Sel1l2
|
SEL1L2 ERAD E3 ligase adaptor subunit |
| chr18_-_399242 | 6.28 |
ENSRNOT00000045926
|
F8
|
coagulation factor VIII |
| chr13_-_102721218 | 6.22 |
ENSRNOT00000005459
|
Marc1
|
mitochondrial amidoxime reducing component 1 |
| chr11_+_52828116 | 6.11 |
ENSRNOT00000035340
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr1_+_147713892 | 5.96 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr18_+_35384743 | 5.90 |
ENSRNOT00000076143
ENSRNOT00000074593 |
LOC100911797
|
serine protease inhibitor Kazal-type 5-like |
| chr14_+_22806132 | 5.86 |
ENSRNOT00000002728
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr16_+_9074033 | 5.82 |
ENSRNOT00000027193
|
Fam170b
|
family with sequence similarity 170, member B |
| chr2_+_205568935 | 5.73 |
ENSRNOT00000025248
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chrX_+_96667863 | 5.72 |
ENSRNOT00000042552
|
RGD1561151
|
similar to hypothetical protein 4932411N23 |
| chr1_+_83163079 | 5.72 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chr19_+_85606 | 5.66 |
ENSRNOT00000015724
|
Ces2e
|
carboxylesterase 2E |
| chr4_+_98337367 | 5.64 |
ENSRNOT00000042165
|
AABR07060872.1
|
|
| chr3_-_134340801 | 5.56 |
ENSRNOT00000085149
|
Sel1l2
|
SEL1L2 ERAD E3 ligase adaptor subunit |
| chr2_-_192288568 | 5.54 |
ENSRNOT00000067346
|
AABR07012291.2
|
|
| chr1_+_83933942 | 5.54 |
ENSRNOT00000068690
|
Cyp2f4
|
cytochrome P450, family 2, subfamily f, polypeptide 4 |
| chr1_-_227441442 | 5.52 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr16_-_6404957 | 5.40 |
ENSRNOT00000048459
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_+_54466280 | 5.40 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr13_-_53870428 | 5.33 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_+_129595192 | 5.31 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr14_-_1505085 | 5.29 |
ENSRNOT00000090361
|
LOC102554799
|
uncharacterized LOC102554799 |
| chr5_-_163167299 | 5.22 |
ENSRNOT00000022478
|
Tnfrsf1b
|
TNF receptor superfamily member 1B |
| chr3_+_55461420 | 5.22 |
ENSRNOT00000073549
|
G6pc2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr2_-_180914940 | 5.10 |
ENSRNOT00000015732
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chrX_+_43293551 | 5.09 |
ENSRNOT00000084562
|
LOC680190
|
hypothetical protein LOC680190 |
| chr2_-_132301073 | 5.05 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr7_-_54823956 | 5.00 |
ENSRNOT00000073180
|
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr1_-_116919269 | 4.95 |
ENSRNOT00000035149
|
AABR07003616.1
|
|
| chr8_-_38549268 | 4.95 |
ENSRNOT00000088001
|
LOC100912026
|
urinary protein 3-like |
| chr2_-_208623314 | 4.92 |
ENSRNOT00000022731
|
Pifo
|
primary cilia formation |
| chr13_+_89919667 | 4.91 |
ENSRNOT00000006196
|
Itln1
|
intelectin 1 |
| chr4_-_162649618 | 4.89 |
ENSRNOT00000050861
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr3_-_153246433 | 4.78 |
ENSRNOT00000067748
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chrX_+_122313470 | 4.77 |
ENSRNOT00000018322
|
RGD1566387
|
similar to hypothetical protein |
| chr1_-_222118459 | 4.73 |
ENSRNOT00000067217
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr1_-_201702963 | 4.72 |
ENSRNOT00000031258
|
LOC499276
|
similar to RIKEN cDNA 1700022C21 |
| chr4_-_163849618 | 4.67 |
ENSRNOT00000086363
ENSRNOT00000077637 |
Ly49si1
|
immunoreceptor Ly49si1 |
| chr4_+_155009479 | 4.52 |
ENSRNOT00000070906
|
AC127013.1
|
|
| chr17_-_57394985 | 4.50 |
ENSRNOT00000019968
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr4_+_96562725 | 4.49 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr16_-_6405117 | 4.48 |
ENSRNOT00000047737
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_-_192381716 | 4.46 |
ENSRNOT00000064950
|
AABR07012291.1
|
|
| chr10_-_17415251 | 4.40 |
ENSRNOT00000005509
|
Efcab9
|
EF-hand calcium binding domain 9 |
| chr1_+_48039909 | 4.38 |
ENSRNOT00000060412
ENSRNOT00000076338 |
Pnldc1
|
PARN like, ribonuclease domain containing 1 |
| chr4_+_165732643 | 4.26 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr4_-_164051812 | 4.26 |
ENSRNOT00000085719
|
AABR07062183.1
|
|
| chr9_-_105282292 | 4.24 |
ENSRNOT00000071358
|
LOC102554121
|
solute carrier organic anion transporter family member 6A1-like |
| chr1_-_169005190 | 4.22 |
ENSRNOT00000043719
|
Hbe1
|
hemoglobin subunit epsilon 1 |
| chr2_+_252452269 | 4.16 |
ENSRNOT00000021970
|
Uox
|
urate oxidase |
| chr1_+_201672528 | 4.04 |
ENSRNOT00000093490
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr6_-_86223052 | 4.03 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr3_-_110021149 | 4.02 |
ENSRNOT00000007808
|
Fsip1
|
fibrous sheath interacting protein 1 |
| chr1_+_55219773 | 4.01 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr5_+_117586103 | 4.00 |
ENSRNOT00000084640
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr17_+_57676569 | 3.97 |
ENSRNOT00000072747
|
RGD1564347
|
similar to isopentenyl diphosphate delta-isomerase type 2 |
| chr2_+_127538659 | 3.96 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr17_-_42639922 | 3.91 |
ENSRNOT00000081105
|
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr1_+_214183724 | 3.90 |
ENSRNOT00000091150
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr1_+_77967755 | 3.87 |
ENSRNOT00000058241
|
Zfp541
|
zinc finger protein 541 |
| chr10_-_64550145 | 3.87 |
ENSRNOT00000050232
|
Nxn
|
nucleoredoxin |
| chr13_+_75177965 | 3.83 |
ENSRNOT00000007321
|
Sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chrX_+_18163358 | 3.81 |
ENSRNOT00000045517
|
LOC367746
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr2_+_147496229 | 3.78 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr1_-_220136470 | 3.70 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chr19_+_37090056 | 3.68 |
ENSRNOT00000019169
|
Ces4a
|
carboxylesterase 4A |
| chrX_+_97074710 | 3.68 |
ENSRNOT00000044379
|
RGD1561230
|
similar to RIKEN cDNA 4921511C20 gene |
| chr4_+_70572942 | 3.67 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr4_-_163445302 | 3.64 |
ENSRNOT00000087106
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chrX_-_63999622 | 3.64 |
ENSRNOT00000090902
|
LOC103694487
|
protein gar2-like |
| chr11_-_14304603 | 3.63 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr13_-_80819218 | 3.62 |
ENSRNOT00000072922
|
Fmo6
|
flavin containing monooxygenase 6 |
| chr19_+_15081590 | 3.60 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr12_+_13284532 | 3.59 |
ENSRNOT00000084045
|
Zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr19_-_22632071 | 3.59 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr7_-_107223047 | 3.53 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr3_+_73335149 | 3.49 |
ENSRNOT00000042537
|
Olr470
|
olfactory receptor 470 |
| chr14_+_84282073 | 3.44 |
ENSRNOT00000078271
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr11_+_86094567 | 3.44 |
ENSRNOT00000086514
|
LOC100361706
|
lambda-chain C1-region-like |
| chr6_-_141472746 | 3.41 |
ENSRNOT00000048010
|
AABR07065792.2
|
|
| chr6_+_127743971 | 3.37 |
ENSRNOT00000013045
|
Serpina4
|
serpin family A member 4 |
| chr9_+_77320726 | 3.36 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr11_+_45751812 | 3.35 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr12_+_30606161 | 3.35 |
ENSRNOT00000093601
|
Sept14
|
septin 14 |
| chr1_-_201894775 | 3.33 |
ENSRNOT00000050519
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chrY_+_184603 | 3.31 |
ENSRNOT00000077708
|
LOC103694540
|
zinc finger Y-chromosomal protein 2 |
| chr14_+_97686121 | 3.31 |
ENSRNOT00000074877
|
Pom121l12
|
POM121 transmembrane nucleoporin-like 12 |
| chr1_-_227506822 | 3.26 |
ENSRNOT00000091506
|
Ms4a7
|
membrane spanning 4-domains A7 |
| chr2_+_154921999 | 3.25 |
ENSRNOT00000057620
|
LOC691044
|
similar to GTPase activating protein testicular GAP1 |
| chrX_-_51792597 | 3.25 |
ENSRNOT00000072727
|
LOC102549011
|
titin-like |
| chr16_-_31301880 | 3.24 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr8_+_106503504 | 3.22 |
ENSRNOT00000018755
|
Rbp2
|
retinol binding protein 2 |
| chr11_+_85561460 | 3.22 |
ENSRNOT00000075455
|
AABR07072262.1
|
|
| chr19_-_49623758 | 3.21 |
ENSRNOT00000071130
|
Pkd1l2
|
polycystic kidney disease 1-like 2 |
| chr5_+_117698764 | 3.18 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr7_+_49385705 | 3.17 |
ENSRNOT00000006083
|
Lin7a
|
lin-7 homolog A, crumbs cell polarity complex component |
| chr14_+_22517774 | 3.15 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chrX_+_57866406 | 3.12 |
ENSRNOT00000076221
|
Ppp4r3c
|
protein phosphatase 4 regulatory subunit 3C |
| chrX_+_57870445 | 3.11 |
ENSRNOT00000065038
|
Ppp4r3c
|
protein phosphatase 4 regulatory subunit 3C |
| chr13_-_27192592 | 3.11 |
ENSRNOT00000040021
|
Serpinb3
|
serpin family B member 3 |
| chr18_-_15688117 | 3.10 |
ENSRNOT00000022584
|
Dsg3
|
desmoglein 3 |
| chr4_-_176026133 | 3.07 |
ENSRNOT00000043374
ENSRNOT00000046598 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr15_+_31164401 | 3.02 |
ENSRNOT00000084286
|
AABR07017798.1
|
|
| chr15_-_38709984 | 2.98 |
ENSRNOT00000074738
|
Phf11b
|
PHD finger protein 11B |
| chr20_-_5166252 | 2.97 |
ENSRNOT00000001138
|
Aif1
|
allograft inflammatory factor 1 |
| chr1_-_73682247 | 2.97 |
ENSRNOT00000079498
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr4_-_78759408 | 2.95 |
ENSRNOT00000050994
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr16_+_72010106 | 2.95 |
ENSRNOT00000058330
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr1_-_227457629 | 2.95 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr8_-_61917125 | 2.92 |
ENSRNOT00000085049
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr17_-_32076181 | 2.92 |
ENSRNOT00000074842
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr15_+_61826937 | 2.91 |
ENSRNOT00000084005
ENSRNOT00000079417 |
Elf1
|
E74-like factor 1 |
| chr4_-_50693869 | 2.89 |
ENSRNOT00000064228
|
Rnf148
|
ring finger protein 148 |
| chr2_-_29248174 | 2.89 |
ENSRNOT00000071167
|
LOC684444
|
similar to HIStone family member (his-41) |
| chr2_+_192463203 | 2.88 |
ENSRNOT00000081253
|
LOC102552651
|
small proline-rich protein 2H-like |
| chr19_+_487723 | 2.86 |
ENSRNOT00000061734
|
Ces2j
|
carboxylesterase 2J |
| chr1_+_256786124 | 2.85 |
ENSRNOT00000034563
|
Ffar4
|
free fatty acid receptor 4 |
| chr19_+_26022849 | 2.85 |
ENSRNOT00000014887
|
Dnase2
|
deoxyribonuclease 2, lysosomal |
| chrX_-_24120987 | 2.85 |
ENSRNOT00000030377
|
LOC685762
|
hypothetical protein LOC685762 |
| chr15_+_34328851 | 2.84 |
ENSRNOT00000026819
|
Tssk4
|
testis-specific serine kinase 4 |
| chr10_+_59566223 | 2.84 |
ENSRNOT00000024068
ENSRNOT00000088245 |
P2rx1
|
purinergic receptor P2X 1 |
| chr2_+_166856784 | 2.84 |
ENSRNOT00000035765
|
Otol1
|
otolin 1 |
| chr4_-_164899041 | 2.83 |
ENSRNOT00000090316
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr12_-_46889082 | 2.81 |
ENSRNOT00000001525
|
Pla2g1b
|
phospholipase A2 group IB |
| chr10_-_88122233 | 2.81 |
ENSRNOT00000083895
ENSRNOT00000005285 |
Krt14
|
keratin 14 |
| chr3_-_78790691 | 2.80 |
ENSRNOT00000008750
|
Olr724
|
olfactory receptor 724 |
| chr8_-_85720790 | 2.80 |
ENSRNOT00000071049
|
Ooep
|
oocyte expressed protein |
| chr1_-_54854353 | 2.79 |
ENSRNOT00000072895
|
Smok2a
|
sperm motility kinase 2A |
| chr14_+_1318157 | 2.77 |
ENSRNOT00000061889
|
Vom2r67
|
vomeronasal 2 receptor, 67 |
| chr7_-_9174330 | 2.73 |
ENSRNOT00000051063
|
Olr1060
|
olfactory receptor 1060 |
| chr11_-_79703736 | 2.71 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr14_+_39368530 | 2.71 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr2_-_192481273 | 2.70 |
ENSRNOT00000012135
|
Sprr2d
|
small proline-rich protein 2D |
| chr1_+_169951913 | 2.70 |
ENSRNOT00000040861
|
Olr189
|
olfactory receptor 189 |
| chr1_-_150101177 | 2.70 |
ENSRNOT00000042931
|
Olr24
|
olfactory receptor 24 |
| chr10_-_109840047 | 2.69 |
ENSRNOT00000054947
|
Notum
|
NOTUM, palmitoleoyl-protein carboxylesterase |
| chr2_-_192780631 | 2.69 |
ENSRNOT00000012371
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr10_+_53781239 | 2.66 |
ENSRNOT00000082871
|
Myh2
|
myosin heavy chain 2 |
| chr20_-_1109027 | 2.64 |
ENSRNOT00000051315
|
Olr1696
|
olfactory receptor 1696 |
| chr3_+_1413671 | 2.63 |
ENSRNOT00000007675
|
Il1f10
|
interleukin 1 family member 10 |
| chr20_-_5166448 | 2.62 |
ENSRNOT00000076331
|
Aif1
|
allograft inflammatory factor 1 |
| chr11_-_87921679 | 2.62 |
ENSRNOT00000084973
|
Serpind1
|
serpin family D member 1 |
| chr20_-_10013559 | 2.62 |
ENSRNOT00000091623
|
Rsph1
|
radial spoke head 1 homolog |
| chr17_-_51912496 | 2.58 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr1_+_148240504 | 2.57 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr1_+_169919411 | 2.57 |
ENSRNOT00000050034
|
Olr186
|
olfactory receptor 186 |
| chr9_-_54484533 | 2.56 |
ENSRNOT00000083514
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr16_+_69022792 | 2.55 |
ENSRNOT00000035985
|
Got1l1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr17_+_43661222 | 2.53 |
ENSRNOT00000022881
ENSRNOT00000022809 ENSRNOT00000022810 |
Hfe
|
hemochromatosis |
| chr9_-_32868371 | 2.52 |
ENSRNOT00000038369
|
LOC689725
|
similar to chromosome 9 open reading frame 79 |
| chrX_+_118742313 | 2.52 |
ENSRNOT00000045110
|
AABR07041078.1
|
|
| chr13_-_91776397 | 2.49 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr9_+_93030714 | 2.45 |
ENSRNOT00000023581
|
Spata3
|
spermatogenesis associated 3 |
| chr1_-_48559162 | 2.44 |
ENSRNOT00000080352
|
Plg
|
plasminogen |
| chr16_-_6404578 | 2.43 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr14_+_85871597 | 2.43 |
ENSRNOT00000079671
|
Ankrd36
|
ankyrin repeat domain 36 |
| chr17_+_84881414 | 2.43 |
ENSRNOT00000034157
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr11_-_29638922 | 2.42 |
ENSRNOT00000048888
|
LOC100359671
|
ribosomal protein L29-like |
| chr1_-_13341952 | 2.41 |
ENSRNOT00000079531
|
NEWGENE_2319083
|
epithelial cell transforming 2 like |
| chr6_+_36941596 | 2.41 |
ENSRNOT00000083383
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr1_-_149859479 | 2.39 |
ENSRNOT00000052199
|
LOC100910009
|
olfactory receptor 14A2-like |
| chr4_+_78382287 | 2.38 |
ENSRNOT00000084927
|
Gimap5
|
GTPase, IMAP family member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx6_Irx2_Irx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 25.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 4.9 | 14.6 | GO:0046340 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 2.1 | 57.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.9 | 5.6 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 1.9 | 3.7 | GO:1904023 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 1.8 | 5.4 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 1.8 | 8.9 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 1.7 | 10.2 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 1.7 | 10.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 1.6 | 6.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.6 | 6.2 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 1.5 | 10.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.4 | 9.6 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) oxidative stress-induced premature senescence(GO:0090403) |
| 1.4 | 12.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.2 | 7.2 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 1.2 | 4.8 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 1.1 | 3.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.1 | 5.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.0 | 3.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.0 | 8.1 | GO:0015679 | plasma membrane copper ion transport(GO:0015679) |
| 1.0 | 5.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.9 | 2.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.9 | 2.8 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.9 | 2.7 | GO:2000502 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.9 | 5.2 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.9 | 2.6 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.8 | 2.5 | GO:0034759 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.8 | 1.7 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.8 | 3.3 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.8 | 3.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.8 | 2.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.7 | 2.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.7 | 5.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.7 | 1.4 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.7 | 7.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.7 | 4.9 | GO:0009624 | response to nematode(GO:0009624) |
| 0.7 | 3.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.6 | 2.6 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.6 | 1.9 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.6 | 3.5 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.6 | 1.7 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.5 | 1.6 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.5 | 2.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 3.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 | 1.6 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 0.5 | 10.0 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.5 | 1.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.5 | 2.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 1.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.5 | 16.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.4 | 1.3 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.4 | 3.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.4 | 4.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.4 | 3.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 2.3 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.4 | 1.5 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.4 | 1.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.4 | 1.5 | GO:0060460 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.4 | 0.7 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.4 | 1.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 7.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 1.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.4 | 1.5 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 1.4 | GO:0035697 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) diapedesis(GO:0050904) cellular response to vitamin K(GO:0071307) |
| 0.4 | 1.1 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.3 | 1.0 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.3 | 4.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 5.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.3 | 2.9 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 4.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 12.4 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.3 | 2.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 5.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 1.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.3 | 1.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 0.6 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.3 | 2.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.3 | 4.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.3 | 2.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 2.2 | GO:0006067 | ethanol metabolic process(GO:0006067) |
| 0.3 | 0.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 2.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 4.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 0.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.3 | 2.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.3 | 2.4 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.3 | 1.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.3 | 1.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.9 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 1.4 | GO:2001184 | toll-like receptor 7 signaling pathway(GO:0034154) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 0.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 0.7 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 1.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.2 | 1.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.2 | 1.7 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 4.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.2 | 8.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 2.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 0.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 4.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 2.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.2 | 1.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.2 | 5.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 2.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 0.6 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.2 | 0.9 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 0.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.2 | 1.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 0.9 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.2 | 1.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 1.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.5 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.2 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.5 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 0.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.4 | GO:0002638 | negative regulation of immunoglobulin production(GO:0002638) |
| 0.1 | 0.3 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 | 0.4 | GO:1903116 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of actin filament-based movement(GO:1903116) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 1.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 1.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 1.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.4 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.9 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 3.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.8 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 3.8 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 1.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 1.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 2.0 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.6 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 1.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 1.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.6 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 2.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 6.4 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.5 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.4 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 2.9 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.1 | 1.6 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 0.9 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 1.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.6 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.1 | 9.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 1.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.0 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.3 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 0.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.2 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 3.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0070055 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.1 | 1.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.6 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.1 | 1.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 2.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 2.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.2 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.1 | 0.5 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.3 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.1 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.6 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 1.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.1 | 3.5 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.1 | 2.5 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 6.8 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 1.0 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.2 | GO:0007308 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.1 | 3.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 3.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 3.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 1.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.4 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 1.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 9.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 4.9 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 | 0.9 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.1 | GO:0042108 | positive regulation of cytokine biosynthetic process(GO:0042108) |
| 0.0 | 1.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 2.0 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.0 | 0.3 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 2.8 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.8 | GO:0010596 | negative regulation of endothelial cell migration(GO:0010596) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.9 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.5 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 1.4 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.5 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.3 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 1.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.5 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 13.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 25.9 | GO:0098536 | deuterosome(GO:0098536) |
| 1.7 | 10.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.6 | 14.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.2 | 7.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.1 | 4.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.0 | 12.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.0 | 5.8 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.9 | 2.6 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.8 | 2.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.8 | 21.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.7 | 3.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.7 | 4.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 3.9 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.6 | 2.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 10.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 9.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.5 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.5 | 2.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.4 | 3.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.4 | 7.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.4 | 2.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 2.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 4.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 6.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 5.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 3.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 2.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 1.3 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.3 | 1.9 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.3 | 2.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 0.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 5.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 2.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 5.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 23.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 4.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.9 | GO:0031262 | condensed nuclear chromosome outer kinetochore(GO:0000942) Ndc80 complex(GO:0031262) |
| 0.2 | 1.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 2.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 1.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 7.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 1.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.5 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 10.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 5.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.2 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 10.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 4.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 9.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 50.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 4.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 3.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 6.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 35.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.3 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.2 | 8.9 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 2.1 | 12.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 2.0 | 10.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.9 | 57.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.9 | 13.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.7 | 5.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.6 | 4.9 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.6 | 6.2 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 1.4 | 5.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.4 | 4.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 1.4 | 4.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.3 | 5.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 1.2 | 8.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 1.2 | 7.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.1 | 5.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.0 | 3.9 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.9 | 2.6 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.8 | 2.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.7 | 2.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.7 | 10.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.7 | 8.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.6 | 2.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 3.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.6 | 19.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 3.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.6 | 1.8 | GO:0005503 | all-trans retinal binding(GO:0005503) benzaldehyde dehydrogenase activity(GO:0019115) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.6 | 4.8 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.6 | 4.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.6 | 2.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 3.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.5 | 6.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 2.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.5 | 1.5 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.5 | 1.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 1.8 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.4 | 14.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 9.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 1.2 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.4 | 11.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.4 | 3.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.4 | 1.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.4 | 1.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.4 | 1.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 1.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 1.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 3.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 6.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 1.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 4.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 5.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.3 | 1.0 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.3 | 1.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.3 | 2.8 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 1.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 2.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 0.9 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 0.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 0.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 3.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 0.8 | GO:0017113 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.3 | 0.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 0.8 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.3 | 10.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 4.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 1.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 8.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 2.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 3.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 5.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 9.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 3.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 2.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.7 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.2 | 1.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 24.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 1.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 3.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 2.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 4.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 2.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 0.5 | GO:0003921 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.2 | 3.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 2.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 3.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.2 | 18.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 1.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 2.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 10.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 3.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 1.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 6.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.6 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 1.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 2.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.6 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 3.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 4.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.8 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 19.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 10.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 4.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 2.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 1.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 4.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.7 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 5.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0031763 | galanin receptor binding(GO:0031763) type 2 galanin receptor binding(GO:0031765) type 3 galanin receptor binding(GO:0031766) |
| 0.0 | 0.8 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 15.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 3.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 6.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.7 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 2.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 5.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 3.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 3.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 2.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 8.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 5.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 35.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 13.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 3.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 5.7 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 14.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 16.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 5.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 5.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 7.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 3.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 5.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 5.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 8.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 4.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 3.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 2.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 10.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 2.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 9.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 4.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 5.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 5.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.1 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.1 | 3.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 3.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |