Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Irx5
Z-value: 0.90
Transcription factors associated with Irx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx5
|
ENSRNOG00000011180 | iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx5 | rn6_v1_chr19_+_16415636_16415636 | -0.18 | 1.3e-03 | Click! |
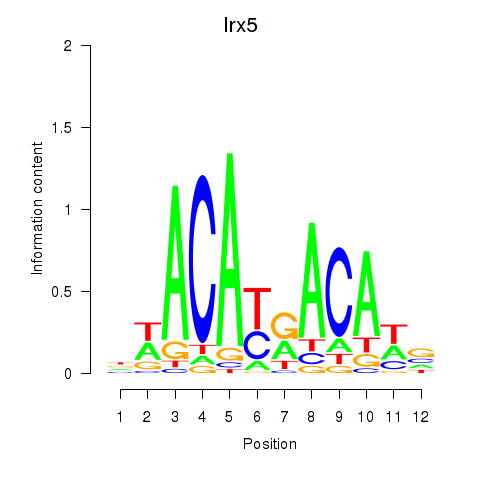
Activity profile of Irx5 motif
Sorted Z-values of Irx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_189328246 | 42.63 |
ENSRNOT00000084260
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr7_-_123655896 | 42.09 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr12_-_29743705 | 28.63 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr2_-_235161263 | 28.39 |
ENSRNOT00000080235
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr1_-_259287684 | 28.35 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr13_-_56763981 | 27.76 |
ENSRNOT00000087916
|
LOC100361907
|
complement factor H-related protein B |
| chr3_+_40038336 | 27.64 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr7_-_123621102 | 27.22 |
ENSRNOT00000046024
|
Cyp2d5
|
cytochrome P450, family 2, subfamily d, polypeptide 5 |
| chr7_-_123630045 | 25.65 |
ENSRNOT00000050002
|
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr7_-_123638702 | 24.55 |
ENSRNOT00000082473
ENSRNOT00000044470 ENSRNOT00000092017 |
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr5_-_166116516 | 22.69 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr17_-_10439691 | 20.74 |
ENSRNOT00000024151
|
Cdhr2
|
cadherin-related family member 2 |
| chr8_+_33514042 | 20.15 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr3_+_134440195 | 19.41 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chr16_+_50181316 | 17.51 |
ENSRNOT00000077662
|
F11
|
coagulation factor XI |
| chr2_+_251200686 | 15.98 |
ENSRNOT00000019210
|
Col24a1
|
collagen type XXIV alpha 1 chain |
| chr1_+_80141630 | 15.04 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr4_-_52350624 | 14.68 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
| chr7_+_26522314 | 13.16 |
ENSRNOT00000011572
|
Slc41a2
|
solute carrier family 41 member 2 |
| chr8_+_5676665 | 12.14 |
ENSRNOT00000012310
|
Mmp3
|
matrix metallopeptidase 3 |
| chr14_-_46153212 | 11.82 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr8_+_57936650 | 11.62 |
ENSRNOT00000089686
|
Exph5
|
exophilin 5 |
| chr2_+_228544418 | 10.09 |
ENSRNOT00000013030
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr3_+_112916217 | 9.56 |
ENSRNOT00000030065
|
Tmem62
|
transmembrane protein 62 |
| chr1_+_268189277 | 9.14 |
ENSRNOT00000065001
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr7_+_139685573 | 8.41 |
ENSRNOT00000088376
|
Pfkm
|
phosphofructokinase, muscle |
| chr6_+_107999846 | 8.25 |
ENSRNOT00000058095
|
Ptgr2
|
prostaglandin reductase 2 |
| chr1_-_100969560 | 8.06 |
ENSRNOT00000035908
|
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr1_+_261497059 | 7.60 |
ENSRNOT00000020390
|
Golga7b
|
golgin A7 family, member B |
| chr8_+_42511520 | 7.59 |
ENSRNOT00000042974
|
Olr1259
|
olfactory receptor 1259 |
| chr16_+_25773602 | 7.42 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr20_-_422464 | 7.39 |
ENSRNOT00000051646
|
Olr1673
|
olfactory receptor 1673 |
| chr9_+_65013862 | 7.18 |
ENSRNOT00000092906
ENSRNOT00000081146 |
Aox3
|
aldehyde oxidase 3 |
| chr13_+_57243877 | 7.11 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr10_-_12818137 | 7.08 |
ENSRNOT00000060972
|
Olr1382
|
olfactory receptor 1382 |
| chr7_-_124999137 | 7.05 |
ENSRNOT00000039228
|
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr2_+_208738132 | 6.64 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chr18_-_28017925 | 6.60 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr4_+_29978739 | 6.58 |
ENSRNOT00000011756
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr2_+_104744461 | 6.57 |
ENSRNOT00000016083
ENSRNOT00000082627 |
Cp
|
ceruloplasmin |
| chr5_+_122390522 | 6.47 |
ENSRNOT00000064567
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr10_-_36336628 | 6.38 |
ENSRNOT00000030035
|
Zfp879
|
zinc finger protein 879 |
| chr11_-_45688560 | 6.01 |
ENSRNOT00000081508
|
Olr1536
|
olfactory receptor 1536 |
| chr10_+_55997614 | 5.85 |
ENSRNOT00000013071
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr3_+_77018186 | 5.57 |
ENSRNOT00000074443
|
Olr646
|
olfactory receptor 646 |
| chr17_+_76359022 | 5.36 |
ENSRNOT00000039621
ENSRNOT00000092481 |
Sec61a2
|
Sec61 translocon alpha 2 subunit |
| chr4_-_142970245 | 5.13 |
ENSRNOT00000067815
|
AC112018.1
|
|
| chr3_-_78571715 | 4.83 |
ENSRNOT00000044992
|
Olr712
|
olfactory receptor 712 |
| chrM_+_3904 | 4.83 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr3_-_103217528 | 4.76 |
ENSRNOT00000074456
|
Olr784
|
olfactory receptor 784 |
| chr4_+_21462779 | 4.70 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr1_-_276228574 | 4.65 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr4_-_103403976 | 4.58 |
ENSRNOT00000088153
|
AABR07061057.3
|
|
| chrX_-_119162518 | 4.48 |
ENSRNOT00000073176
|
LOC103694506
|
ubiquitin-conjugating enzyme E2 W |
| chr1_+_68528976 | 4.46 |
ENSRNOT00000072292
|
Vom1r27
|
vomeronasal 1 receptor 27 |
| chr10_-_52290657 | 4.32 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr1_+_75194565 | 4.28 |
ENSRNOT00000050604
|
Vom1r60
|
vomeronasal 1 receptor 60 |
| chr1_-_60634546 | 4.17 |
ENSRNOT00000051451
|
Vom1r15
|
vomeronasal 1 receptor 15 |
| chrX_-_37003642 | 4.09 |
ENSRNOT00000040770
ENSRNOT00000058834 ENSRNOT00000058833 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr1_+_168196862 | 4.06 |
ENSRNOT00000021006
|
Olr74
|
olfactory receptor 74 |
| chr1_+_229349133 | 4.02 |
ENSRNOT00000075498
|
LOC100910906
|
serine protease inhibitor Kazal-type 14-like |
| chr1_+_201901342 | 4.01 |
ENSRNOT00000036657
|
RGD1559891
|
similar to synaptonemal complex protein 3 |
| chr2_-_259382765 | 3.99 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr10_-_12674077 | 3.85 |
ENSRNOT00000041353
|
Olr1376
|
olfactory receptor 1376 |
| chr19_+_51317425 | 3.83 |
ENSRNOT00000019298
|
Cdh13
|
cadherin 13 |
| chr1_+_157920786 | 3.77 |
ENSRNOT00000014284
|
Fam181b
|
family with sequence similarity 181, member B |
| chr3_-_78595778 | 3.73 |
ENSRNOT00000084255
|
Olr715
|
olfactory receptor 715 |
| chr1_-_230687157 | 3.71 |
ENSRNOT00000048193
|
Olr380
|
olfactory receptor 380 |
| chr1_+_167786337 | 3.67 |
ENSRNOT00000024945
|
Olr48
|
olfactory receptor 48 |
| chr2_-_149754776 | 3.63 |
ENSRNOT00000035057
|
RGD1559622
|
similar to hypothetical protein C130079G13 |
| chr5_+_69305190 | 3.55 |
ENSRNOT00000074390
|
LOC108348075
|
olfactory receptor 13C3-like |
| chr3_-_2845593 | 3.54 |
ENSRNOT00000067479
|
Tmem141
|
transmembrane protein 141 |
| chr3_-_73543088 | 3.51 |
ENSRNOT00000012912
|
Olr485
|
olfactory receptor 485 |
| chr9_-_92524739 | 3.38 |
ENSRNOT00000089889
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr4_+_72603242 | 3.34 |
ENSRNOT00000007229
|
Olr818
|
olfactory receptor 818 |
| chr1_-_103811148 | 3.27 |
ENSRNOT00000030162
|
Mrgprx2l
|
MAS-related G protein-coupled receptor, member X2-like |
| chr7_-_6803318 | 3.21 |
ENSRNOT00000084203
|
Olr954
|
olfactory receptor 954 |
| chr3_-_78790691 | 3.08 |
ENSRNOT00000008750
|
Olr724
|
olfactory receptor 724 |
| chr3_+_93968855 | 2.99 |
ENSRNOT00000014478
|
Fbxo3
|
F-box protein 3 |
| chrX_+_159560292 | 2.98 |
ENSRNOT00000087250
|
Vgll1
|
vestigial like family member 1 |
| chr7_+_9279620 | 2.79 |
ENSRNOT00000011225
|
Olr1064
|
olfactory receptor 1064 |
| chr8_-_114013623 | 2.78 |
ENSRNOT00000018175
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr3_-_103128243 | 2.72 |
ENSRNOT00000046310
|
Olr779
|
olfactory receptor 779 |
| chr1_+_168341595 | 2.60 |
ENSRNOT00000048411
|
Olr84
|
olfactory receptor 84 |
| chr1_-_168611670 | 2.59 |
ENSRNOT00000021273
|
Olr106
|
olfactory receptor 106 |
| chr7_-_5106708 | 2.49 |
ENSRNOT00000046001
|
Olr892
|
olfactory receptor 892 |
| chr7_-_15856676 | 2.48 |
ENSRNOT00000086553
|
AABR07055919.1
|
|
| chr3_-_78409263 | 2.44 |
ENSRNOT00000041001
|
Olr704
|
olfactory receptor 704 |
| chr5_-_69277783 | 2.36 |
ENSRNOT00000046638
|
Olr847
|
olfactory receptor 847 |
| chr1_-_149175205 | 2.29 |
ENSRNOT00000089091
ENSRNOT00000045611 ENSRNOT00000089015 |
Vom2r42
|
vomeronasal 2 receptor, 42 |
| chr1_+_225690756 | 2.25 |
ENSRNOT00000027498
|
Psbpc2
|
prostatic steroid-binding protein C2 |
| chr3_-_73580594 | 2.22 |
ENSRNOT00000012929
|
Olr488
|
olfactory receptor 488 |
| chr3_-_78663607 | 2.04 |
ENSRNOT00000073134
|
Olr718
|
olfactory receptor 718 |
| chr3_-_77792014 | 1.99 |
ENSRNOT00000091085
|
Olr671
|
olfactory receptor 671 |
| chrX_-_152110010 | 1.98 |
ENSRNOT00000044666
|
LOC100362263
|
melanoma antigen family A, 5-like |
| chr1_-_147272006 | 1.95 |
ENSRNOT00000086862
|
AABR07004539.1
|
|
| chr1_+_60117804 | 1.90 |
ENSRNOT00000080437
|
Vom1r8
|
vomeronasal 1 receptor 8 |
| chr3_+_75768699 | 1.79 |
ENSRNOT00000071912
|
AABR07052798.1
|
|
| chr17_-_42817037 | 1.73 |
ENSRNOT00000023166
|
Prl3c1
|
Prolactin family 3, subfamily c, member 1 |
| chr3_-_78886096 | 1.72 |
ENSRNOT00000008864
|
Olr729
|
olfactory receptor 729 |
| chr7_+_47928060 | 1.71 |
ENSRNOT00000005740
|
Ccdc59
|
coiled-coil domain containing 59 |
| chr4_+_87312766 | 1.69 |
ENSRNOT00000052126
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr20_-_11498277 | 1.69 |
ENSRNOT00000046763
|
AABR07044581.1
|
|
| chr3_+_75834770 | 1.68 |
ENSRNOT00000043371
|
Olr582
|
olfactory receptor 582 |
| chr1_+_72230038 | 1.58 |
ENSRNOT00000058997
|
Vom1r38
|
vomeronasal 1 receptor 38 |
| chr4_+_157057601 | 1.54 |
ENSRNOT00000040592
|
Zfp42l
|
Zinc finger protein 42-like |
| chr7_-_3707226 | 1.43 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr3_-_78555530 | 1.42 |
ENSRNOT00000089672
|
Olr711
|
olfactory receptor 711 |
| chr5_+_108935189 | 1.31 |
ENSRNOT00000071169
|
AABR07049184.1
|
|
| chr1_+_168403613 | 1.30 |
ENSRNOT00000079028
|
Olr88
|
olfactory receptor 88 |
| chr1_+_168411463 | 1.28 |
ENSRNOT00000072397
|
Olr89
|
olfactory receptor 89 |
| chr1_+_230604386 | 1.25 |
ENSRNOT00000040608
|
Olr375
|
olfactory receptor 375 |
| chr3_+_75437833 | 1.24 |
ENSRNOT00000042246
|
Olr561
|
olfactory receptor 561 |
| chr3_-_76696107 | 1.23 |
ENSRNOT00000044692
|
Olr629
|
olfactory receptor 629 |
| chr11_-_43594659 | 1.22 |
ENSRNOT00000086976
|
Olr1555
|
olfactory receptor 1555 |
| chr11_-_70211701 | 1.05 |
ENSRNOT00000076114
|
Muc13
|
mucin 13, cell surface associated |
| chr20_+_734642 | 0.99 |
ENSRNOT00000073543
|
Olr1688
|
olfactory receptor 1688 |
| chr7_-_47928674 | 0.96 |
ENSRNOT00000038144
|
Mettl25
|
methyltransferase like 25 |
| chr10_+_61160125 | 0.95 |
ENSRNOT00000074715
|
Olr1519
|
olfactory receptor 1519 |
| chr18_-_26658892 | 0.91 |
ENSRNOT00000038247
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr1_-_169770188 | 0.88 |
ENSRNOT00000072602
|
Olr165
|
olfactory receptor 165 |
| chr3_-_74790822 | 0.86 |
ENSRNOT00000045888
|
LOC100912505
|
olfactory receptor 8K3-like |
| chr8_+_25246292 | 0.86 |
ENSRNOT00000021625
|
Npsr1
|
neuropeptide S receptor 1 |
| chr8_-_50246656 | 0.78 |
ENSRNOT00000024240
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr1_-_168015148 | 0.72 |
ENSRNOT00000020691
|
Olr50
|
olfactory receptor 50 |
| chr1_-_66688422 | 0.66 |
ENSRNOT00000082786
|
Vom1r57
|
vomeronasal 1 receptor 57 |
| chr1_-_60281386 | 0.63 |
ENSRNOT00000092184
|
Vom1r10
|
vomeronasal 1 receptor 10 |
| chr8_-_117237229 | 0.63 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr1_-_172030164 | 0.53 |
ENSRNOT00000026807
|
Ovch2
|
ovochymase 2 |
| chr17_+_79749747 | 0.48 |
ENSRNOT00000081743
|
AABR07028665.1
|
|
| chr2_-_220838905 | 0.46 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr7_-_143167772 | 0.37 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr2_+_194557986 | 0.29 |
ENSRNOT00000070843
|
LOC100911679
|
TD and POZ domain-containing protein 2-like |
| chr14_+_1318157 | 0.25 |
ENSRNOT00000061889
|
Vom2r67
|
vomeronasal 2 receptor, 67 |
| chr3_+_76768051 | 0.19 |
ENSRNOT00000047382
|
Olr634
|
olfactory receptor 634 |
| chr1_-_67284864 | 0.14 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr3_-_74906989 | 0.13 |
ENSRNOT00000071001
|
Olr545
|
olfactory receptor 545 |
| chr1_+_75748916 | 0.12 |
ENSRNOT00000083154
ENSRNOT00000089836 |
Bsph1
|
binder of sperm protein homolog 1 |
| chr8_+_19888667 | 0.09 |
ENSRNOT00000078593
|
Zfp317
|
zinc finger protein 317 |
| chr1_-_168310071 | 0.06 |
ENSRNOT00000044675
|
Olr82
|
olfactory receptor 82 |
| chr3_+_75809445 | 0.05 |
ENSRNOT00000049950
|
Olr581
|
olfactory receptor 581 |
| chr1_+_169741497 | 0.02 |
ENSRNOT00000072865
|
Olr164
|
olfactory receptor 164 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 4.5 | 22.7 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 4.2 | 50.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 4.1 | 20.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 2.5 | 17.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 2.1 | 8.4 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 1.7 | 12.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.6 | 97.7 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 1.4 | 7.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 1.2 | 11.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 1.1 | 6.6 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 1.1 | 4.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.0 | 7.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.9 | 2.8 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.9 | 6.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.8 | 6.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.8 | 6.6 | GO:0015679 | plasma membrane copper ion transport(GO:0015679) |
| 0.5 | 9.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.5 | 8.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.5 | 15.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.5 | 27.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.5 | 3.8 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.5 | 42.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.4 | 4.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 8.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.2 | 4.0 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.2 | 4.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.2 | 1.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 4.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.9 | GO:2000293 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 7.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 3.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 4.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 17.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 102.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 9.1 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 7.1 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 13.1 | GO:0070838 | divalent metal ion transport(GO:0070838) |
| 0.0 | 4.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 14.1 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 2.8 | 8.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 2.5 | 22.7 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.6 | 6.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.5 | 5.9 | GO:0031417 | NatC complex(GO:0031417) |
| 1.0 | 28.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 7.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.4 | 6.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 49.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 8.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 4.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 4.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 147.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 4.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 7.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 34.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 4.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 7.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 21.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 15.7 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 185.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 11.6 | GO:0005768 | endosome(GO:0005768) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 42.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 5.0 | 20.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 3.8 | 22.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 3.6 | 147.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 2.8 | 8.3 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 2.5 | 27.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 2.1 | 8.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 2.0 | 8.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.6 | 4.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 1.5 | 17.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.4 | 7.2 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 1.1 | 6.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.0 | 3.8 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.8 | 4.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 6.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 4.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 7.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 2.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.3 | 7.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 16.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 6.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 7.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 4.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 4.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.8 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 15.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 12.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 102.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 5.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.9 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 13.1 | GO:0072509 | divalent inorganic cation transmembrane transporter activity(GO:0072509) |
| 0.1 | 11.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 6.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 4.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 11.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 9.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 3.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 4.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 12.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 8.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 17.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 19.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.6 | 32.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.5 | 20.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.5 | 42.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 16.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 8.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 5.3 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.2 | 4.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 9.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |