Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Irx4
Z-value: 0.38
Transcription factors associated with Irx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx4
|
ENSRNOG00000012720 | iroquois homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx4 | rn6_v1_chr1_-_32643771_32643771 | -0.12 | 3.7e-02 | Click! |
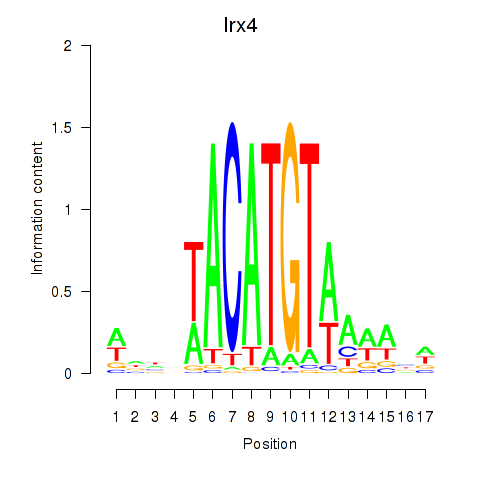
Activity profile of Irx4 motif
Sorted Z-values of Irx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_83103925 | 14.70 |
ENSRNOT00000047540
ENSRNOT00000028196 |
Cyp2b2
|
cytochrome P450, family 2, subfamily b, polypeptide 2 |
| chr2_+_193724248 | 9.90 |
ENSRNOT00000025249
|
Rptn
|
repetin |
| chr4_+_14109864 | 9.46 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr3_-_153246433 | 8.08 |
ENSRNOT00000067748
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr16_-_6404957 | 7.80 |
ENSRNOT00000048459
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr4_-_115015965 | 6.57 |
ENSRNOT00000014603
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr16_-_6405117 | 6.31 |
ENSRNOT00000047737
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr5_-_163167299 | 6.09 |
ENSRNOT00000022478
|
Tnfrsf1b
|
TNF receptor superfamily member 1B |
| chr13_-_67819835 | 6.09 |
ENSRNOT00000093666
|
Hmcn1
|
hemicentin 1 |
| chr11_+_85561460 | 6.06 |
ENSRNOT00000075455
|
AABR07072262.1
|
|
| chr16_-_6404578 | 5.93 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr3_+_1385654 | 4.31 |
ENSRNOT00000091805
ENSRNOT00000007593 |
Il36rn
|
interleukin 36 receptor antagonist |
| chr3_-_46601409 | 3.99 |
ENSRNOT00000079261
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr17_-_51912496 | 3.67 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr18_+_35384743 | 2.86 |
ENSRNOT00000076143
ENSRNOT00000074593 |
LOC100911797
|
serine protease inhibitor Kazal-type 5-like |
| chrX_+_18163358 | 2.56 |
ENSRNOT00000045517
|
LOC367746
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr16_+_72086878 | 2.18 |
ENSRNOT00000023756
ENSRNOT00000078085 |
Adam3a
|
ADAM metallopeptidase domain 3A |
| chr3_+_76044495 | 1.69 |
ENSRNOT00000049471
|
Olr597
|
olfactory receptor 597 |
| chr1_+_169966190 | 1.35 |
ENSRNOT00000023307
|
Olr190
|
olfactory receptor 190 |
| chr7_-_9174330 | 1.34 |
ENSRNOT00000051063
|
Olr1060
|
olfactory receptor 1060 |
| chr3_-_134340801 | 1.16 |
ENSRNOT00000085149
|
Sel1l2
|
SEL1L2 ERAD E3 ligase adaptor subunit |
| chr3_+_75559775 | 1.14 |
ENSRNOT00000035844
|
Pramel6
|
preferentially expressed antigen in melanoma-like 6 |
| chr3_+_73335149 | 0.94 |
ENSRNOT00000042537
|
Olr470
|
olfactory receptor 470 |
| chr13_+_65550673 | 0.90 |
ENSRNOT00000035431
|
LOC689412
|
similar to CG4025-PA |
| chr18_-_60835739 | 0.89 |
ENSRNOT00000039569
|
LOC291543
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr4_-_120559078 | 0.82 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr4_+_82783686 | 0.39 |
ENSRNOT00000041988
|
Tax1bp1
|
Tax1 binding protein 1 |
| chr18_+_29490981 | 0.38 |
ENSRNOT00000032316
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr3_+_75768699 | 0.36 |
ENSRNOT00000071912
|
AABR07052798.1
|
|
| chr1_+_230576298 | 0.21 |
ENSRNOT00000049799
|
Olr374
|
olfactory receptor 374 |
| chr12_+_41385241 | 0.17 |
ENSRNOT00000074974
|
Dtx1
|
deltex E3 ubiquitin ligase 1 |
| chrX_-_120521871 | 0.16 |
ENSRNOT00000080863
|
LOC100362376
|
LRRGT00025-like |
| chr10_-_43895376 | 0.14 |
ENSRNOT00000031811
|
Olr1413
|
olfactory receptor 1413 |
| chr3_+_151688454 | 0.05 |
ENSRNOT00000026856
|
Romo1
|
reactive oxygen species modulator 1 |
| chr20_-_821332 | 0.01 |
ENSRNOT00000040957
|
Olr1692
|
olfactory receptor 1692 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 20.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 2.0 | 8.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 1.3 | 14.7 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.9 | 3.7 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.7 | 4.3 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.6 | 4.0 | GO:0090403 | positive regulation of arachidonic acid secretion(GO:0090238) oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 | 6.6 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.3 | 6.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 20.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.2 | 3.7 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.4 | 9.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 6.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 6.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 13.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 4.0 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 20.0 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.6 | 6.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.0 | 8.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.9 | 3.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 4.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.5 | 14.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 6.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 4.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 16.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 6.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 14.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |