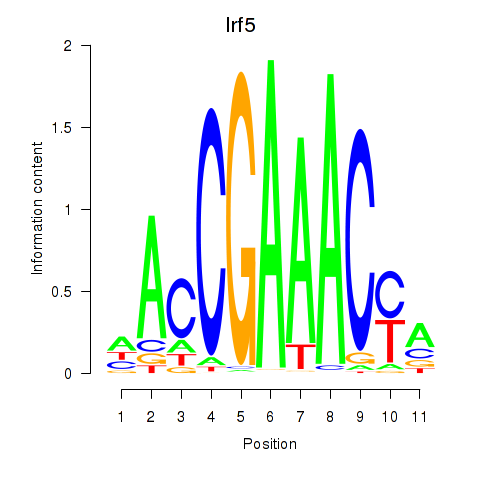
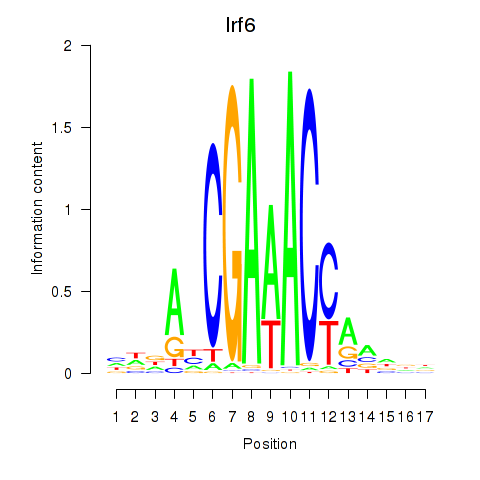
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Irf5_Irf6
Z-value: 0.38
Transcription factors associated with Irf5_Irf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf5
|
ENSRNOG00000007437 | interferon regulatory factor 5 |
|
Irf6
|
ENSRNOG00000005082 | interferon regulatory factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf6 | rn6_v1_chr13_+_111870121_111870121 | 0.62 | 1.6e-35 | Click! |
| Irf5 | rn6_v1_chr4_+_56805132_56805158 | 0.02 | 7.3e-01 | Click! |
Activity profile of Irf5_Irf6 motif
Sorted Z-values of Irf5_Irf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_4542073 | 20.74 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr9_-_4945352 | 19.24 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr10_+_76343847 | 14.25 |
ENSRNOT00000055674
|
Trim25
|
tripartite motif-containing 25 |
| chr5_-_56536772 | 9.59 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr11_+_67757928 | 9.52 |
ENSRNOT00000039215
|
Dtx3l
|
deltex E3 ubiquitin ligase 3L |
| chr2_-_30576591 | 9.36 |
ENSRNOT00000084667
|
Ocln
|
occludin |
| chr2_-_30577218 | 8.45 |
ENSRNOT00000024674
|
Ocln
|
occludin |
| chr4_-_10517832 | 8.38 |
ENSRNOT00000039953
ENSRNOT00000083964 |
Gsap
|
gamma-secretase activating protein |
| chr7_-_101140308 | 6.59 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr5_+_159845774 | 6.41 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr2_+_185393441 | 5.56 |
ENSRNOT00000015802
|
Sh3d19
|
SH3 domain containing 19 |
| chr6_-_51257625 | 5.32 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
| chr1_-_193328371 | 5.28 |
ENSRNOT00000019304
ENSRNOT00000031770 ENSRNOT00000019309 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chr5_+_16526058 | 4.63 |
ENSRNOT00000011130
|
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr5_-_62153762 | 4.62 |
ENSRNOT00000066001
ENSRNOT00000086962 |
Trim14
|
tripartite motif-containing 14 |
| chr15_+_3996048 | 4.43 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr4_+_145427367 | 4.36 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr15_+_3996328 | 4.31 |
ENSRNOT00000088252
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr10_+_13000090 | 4.29 |
ENSRNOT00000004845
|
Cldn6
|
claudin 6 |
| chr20_+_3979035 | 3.71 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr11_+_68105369 | 3.37 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr16_+_59988603 | 3.35 |
ENSRNOT00000015176
|
Cldn23
|
claudin 23 |
| chr10_+_59259955 | 3.24 |
ENSRNOT00000021816
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr5_+_116421894 | 3.13 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chrX_-_79909678 | 2.89 |
ENSRNOT00000050336
|
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr1_+_220948331 | 2.82 |
ENSRNOT00000036185
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr20_-_3978845 | 2.76 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr2_+_240627436 | 2.14 |
ENSRNOT00000066204
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_247988462 | 2.12 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chr10_-_4257868 | 2.05 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr9_-_54327958 | 2.04 |
ENSRNOT00000019465
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr16_-_19349080 | 2.00 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr2_-_127710448 | 1.96 |
ENSRNOT00000093733
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chrX_+_44830849 | 1.82 |
ENSRNOT00000079680
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr13_-_25262469 | 1.75 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr10_-_82963919 | 1.54 |
ENSRNOT00000005807
|
Dlx4
|
distal-less homeobox 4 |
| chr10_-_54467956 | 1.54 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chrX_+_123751089 | 1.47 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr4_-_51726212 | 1.39 |
ENSRNOT00000009261
|
Wasl
|
Wiskott-Aldrich syndrome-like |
| chrX_+_123751293 | 1.11 |
ENSRNOT00000089883
|
Nkap
|
NFKB activating protein |
| chr17_+_36334147 | 1.01 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr1_+_78671121 | 0.97 |
ENSRNOT00000021310
|
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr17_+_6667564 | 0.74 |
ENSRNOT00000025916
|
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr1_-_220938814 | 0.59 |
ENSRNOT00000028081
|
Ovol1
|
ovo like transcriptional repressor 1 |
| chr10_+_46940965 | 0.58 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr3_-_29984201 | 0.54 |
ENSRNOT00000006350
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr8_+_103774358 | 0.47 |
ENSRNOT00000014481
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr13_-_102643223 | 0.36 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr6_-_10592454 | 0.25 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr11_+_43687107 | 0.17 |
ENSRNOT00000048343
|
Olr1558
|
olfactory receptor 1558 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf5_Irf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0009597 | detection of virus(GO:0009597) |
| 1.9 | 20.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.8 | 17.8 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 1.6 | 6.4 | GO:0014028 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) negative regulation of lymphangiogenesis(GO:1901491) |
| 1.5 | 4.6 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.2 | 3.7 | GO:0046967 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 1.0 | 8.7 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.7 | 14.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.7 | 8.4 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.7 | 2.8 | GO:1901423 | response to benzene(GO:1901423) |
| 0.6 | 3.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.5 | 2.0 | GO:0046725 | negative regulation of macrophage fusion(GO:0034240) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.4 | 2.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.4 | 9.5 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.3 | 4.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 5.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.7 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.2 | 1.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 4.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 3.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 5.3 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.2 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.6 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.4 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 2.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 2.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 19.2 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 0.6 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.0 | 2.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 5.3 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.0 | 2.0 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 1.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 2.8 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 2.5 | GO:0045216 | cell-cell junction organization(GO:0045216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.9 | 22.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.5 | 3.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 2.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 3.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 1.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 20.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 18.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 2.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 8.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 6.4 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 16.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 34.7 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 1.5 | 8.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.4 | 14.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 1.2 | 3.7 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.5 | 4.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 4.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 6.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 8.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 5.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 2.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 3.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 9.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 20.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 11.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 12.9 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 2.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 20.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 4.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 6.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 17.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 5.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 2.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 5.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 3.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 4.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 23.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 1.5 | 20.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.3 | 17.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 4.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 8.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 3.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 4.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 5.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.0 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 1.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.8 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.1 | 9.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 5.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |