Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
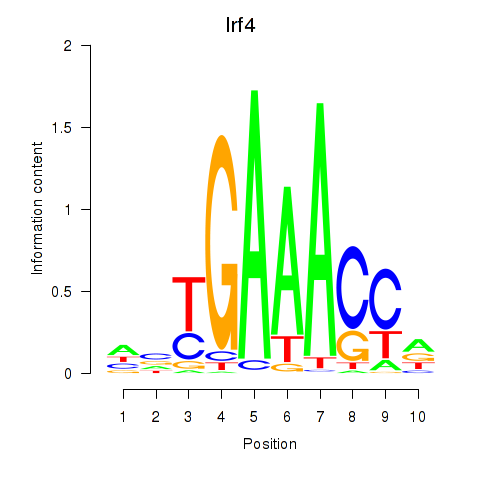
Results for Irf4
Z-value: 0.67
Transcription factors associated with Irf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf4
|
ENSRNOG00000061070 | interferon regulatory factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf4 | rn6_v1_chr17_-_34905117_34905117 | 0.08 | 1.7e-01 | Click! |
Activity profile of Irf4 motif
Sorted Z-values of Irf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_18710492 | 23.38 |
ENSRNOT00000012532
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr1_-_16687817 | 19.41 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr1_+_198744050 | 17.25 |
ENSRNOT00000024404
|
Itgal
|
integrin subunit alpha L |
| chr17_+_72429618 | 16.93 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr11_+_47243342 | 16.49 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr1_+_260289589 | 15.75 |
ENSRNOT00000051058
|
Dntt
|
DNA nucleotidylexotransferase |
| chr10_+_76343847 | 15.46 |
ENSRNOT00000055674
|
Trim25
|
tripartite motif-containing 25 |
| chr11_+_61970976 | 15.25 |
ENSRNOT00000078921
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr10_-_29450644 | 15.06 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr4_-_176679815 | 14.42 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr10_-_88050622 | 14.01 |
ENSRNOT00000019037
|
Krt15
|
keratin 15 |
| chr8_-_124399494 | 13.90 |
ENSRNOT00000037883
|
Tgfbr2
|
transforming growth factor, beta receptor 2 |
| chr9_-_4945352 | 13.74 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr4_-_31730386 | 13.41 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr20_-_4542073 | 12.01 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr1_-_47331412 | 11.78 |
ENSRNOT00000046746
|
Ezr
|
ezrin |
| chr1_-_79930263 | 11.73 |
ENSRNOT00000019219
|
Foxa3
|
forkhead box A3 |
| chr5_-_153924896 | 11.41 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr17_-_69827112 | 11.07 |
ENSRNOT00000023835
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr10_+_34277993 | 10.96 |
ENSRNOT00000055872
ENSRNOT00000003343 |
Ifi47
|
interferon gamma inducible protein 47 |
| chr10_+_31880918 | 10.87 |
ENSRNOT00000059448
|
Timd4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chrX_+_110789269 | 10.53 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr15_+_24141651 | 10.53 |
ENSRNOT00000082304
|
Lgals3
|
galectin 3 |
| chr10_-_89454681 | 10.38 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr2_-_216382244 | 10.05 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr1_-_142183884 | 9.38 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr20_-_35450513 | 9.05 |
ENSRNOT00000087342
|
Man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr11_+_67757928 | 8.90 |
ENSRNOT00000039215
|
Dtx3l
|
deltex E3 ubiquitin ligase 3L |
| chr13_-_99531959 | 8.78 |
ENSRNOT00000005059
|
Wdr26
|
WD repeat domain 26 |
| chr2_-_30576591 | 8.53 |
ENSRNOT00000084667
|
Ocln
|
occludin |
| chr8_-_92942076 | 8.49 |
ENSRNOT00000056937
|
Fam46a
|
family with sequence similarity 46, member A |
| chr20_-_4698718 | 8.36 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr2_+_248178389 | 8.35 |
ENSRNOT00000037339
|
Gbp5
|
guanylate binding protein 5 |
| chr1_+_91857057 | 7.92 |
ENSRNOT00000077993
ENSRNOT00000081465 |
Ankrd27
|
ankyrin repeat domain 27 |
| chr2_-_30577218 | 7.66 |
ENSRNOT00000024674
|
Ocln
|
occludin |
| chr13_+_82438697 | 7.48 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr18_+_24540659 | 7.06 |
ENSRNOT00000061074
|
Ammecr1l
|
AMMECR1 like |
| chr10_-_74724472 | 6.87 |
ENSRNOT00000008846
|
Rad51c
|
RAD51 paralog C |
| chr15_+_34256071 | 6.75 |
ENSRNOT00000025887
|
Psme1
|
proteasome activator subunit 1 |
| chr5_-_4975436 | 6.40 |
ENSRNOT00000062006
|
Xkr9
|
XK related 9 |
| chr20_-_27682861 | 6.34 |
ENSRNOT00000057317
|
Fam26f
|
family with sequence similarity 26, member F |
| chr4_+_161685258 | 6.32 |
ENSRNOT00000008012
ENSRNOT00000008003 |
Foxm1
|
forkhead box M1 |
| chr5_-_62153762 | 6.17 |
ENSRNOT00000066001
ENSRNOT00000086962 |
Trim14
|
tripartite motif-containing 14 |
| chr2_-_30634243 | 5.98 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr4_+_112773938 | 5.77 |
ENSRNOT00000009321
|
Eva1a
|
eva-1 homolog A, regulator of programmed cell death |
| chr15_+_3996328 | 5.72 |
ENSRNOT00000088252
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr3_+_120726906 | 5.70 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr2_-_258997138 | 5.64 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr2_+_185393441 | 5.51 |
ENSRNOT00000015802
|
Sh3d19
|
SH3 domain containing 19 |
| chr5_-_107857320 | 5.48 |
ENSRNOT00000008898
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr1_+_266333440 | 5.47 |
ENSRNOT00000071243
|
Sfxn2
|
sideroflexin 2 |
| chr5_-_59913348 | 4.99 |
ENSRNOT00000018164
|
LOC100359916
|
heterogeneous nuclear ribonucleoprotein K-like |
| chr4_-_157743199 | 4.97 |
ENSRNOT00000038178
|
Tapbpl
|
TAP binding protein-like |
| chr20_-_20713128 | 4.43 |
ENSRNOT00000000784
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_-_40921508 | 4.40 |
ENSRNOT00000026433
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr15_+_3996048 | 4.36 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr7_-_24885775 | 4.14 |
ENSRNOT00000010024
|
Tcp11l2
|
t-complex 11 like 2 |
| chr5_-_56536772 | 3.89 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr6_+_147315328 | 3.88 |
ENSRNOT00000056654
|
Macc1
|
MACC1, MET transcriptional regulator |
| chrX_+_54390733 | 3.85 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr15_-_29291454 | 3.79 |
ENSRNOT00000092212
|
AABR07017617.2
|
|
| chr9_+_4107246 | 3.70 |
ENSRNOT00000078212
|
AABR07066160.1
|
|
| chr3_+_72395218 | 3.60 |
ENSRNOT00000057616
|
Prg3
|
proteoglycan 3, pro eosinophil major basic protein 2 |
| chr17_+_6667564 | 3.51 |
ENSRNOT00000025916
|
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr8_-_55050194 | 3.48 |
ENSRNOT00000013197
|
LOC100151767
|
hypothetical LOC100151767 |
| chr10_-_47775055 | 3.34 |
ENSRNOT00000057864
|
LOC100912585
|
mitogen-activated protein kinase 7-like |
| chr1_+_196095214 | 3.32 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chr20_+_4329811 | 3.17 |
ENSRNOT00000000513
|
Notch4
|
notch 4 |
| chr10_-_34221928 | 3.06 |
ENSRNOT00000045545
|
Irgm
|
immunity-related GTPase M |
| chr10_+_108527740 | 2.77 |
ENSRNOT00000044983
|
Rnf213
|
ring finger protein 213 |
| chr2_-_185852759 | 2.76 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr2_-_211690716 | 2.71 |
ENSRNOT00000027690
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr2_+_150756185 | 2.53 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr1_+_80056755 | 2.53 |
ENSRNOT00000021221
|
Snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr8_+_107826424 | 2.52 |
ENSRNOT00000019769
|
Dbr1
|
debranching RNA lariats 1 |
| chr1_-_214252456 | 2.49 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr7_+_91832988 | 2.44 |
ENSRNOT00000006410
|
Slc30a8
|
solute carrier family 30 member 8 |
| chrX_-_76708878 | 2.39 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr2_-_30633857 | 2.25 |
ENSRNOT00000085488
|
Marveld2
|
MARVEL domain containing 2 |
| chr8_-_40122450 | 2.23 |
ENSRNOT00000039729
|
Tbrg1
|
transforming growth factor beta regulator 1 |
| chr8_+_55050284 | 1.91 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr1_+_13261876 | 1.88 |
ENSRNOT00000090703
|
Reps1
|
RALBP1 associated Eps domain containing 1 |
| chr2_+_95077577 | 1.85 |
ENSRNOT00000046368
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr17_-_32015071 | 1.78 |
ENSRNOT00000070876
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr1_-_173456488 | 1.74 |
ENSRNOT00000044753
|
Olr282
|
olfactory receptor 282 |
| chr15_+_105640097 | 1.60 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chr5_-_107858104 | 1.57 |
ENSRNOT00000092196
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr1_-_242083484 | 1.57 |
ENSRNOT00000065921
|
Tjp2
|
tight junction protein 2 |
| chr1_-_59903339 | 1.55 |
ENSRNOT00000039037
|
Fpr-rs3
|
formyl peptide receptor, related sequence 3 |
| chr8_+_117307339 | 1.51 |
ENSRNOT00000064571
|
Qrich1
|
glutamine-rich 1 |
| chr13_-_102643223 | 1.48 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr6_+_135866739 | 1.29 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr3_+_161178117 | 1.29 |
ENSRNOT00000051826
|
Spint5p
|
serine peptidase inhibitor, Kunitz type 5 |
| chr1_+_168136959 | 1.28 |
ENSRNOT00000020953
|
Olr68
|
olfactory receptor 68 |
| chrX_+_31984612 | 1.17 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr1_-_123064642 | 1.07 |
ENSRNOT00000013551
|
Mkrn3
|
makorin, ring finger protein, 3 |
| chr1_-_198008893 | 0.95 |
ENSRNOT00000025950
|
Il27
|
interleukin 27 |
| chr4_-_10269748 | 0.95 |
ENSRNOT00000074662
|
Fam185a
|
family with sequence similarity 185, member A |
| chr12_-_9998779 | 0.94 |
ENSRNOT00000001265
|
Rpl21
|
ribosomal protein L21 |
| chr5_-_69752314 | 0.93 |
ENSRNOT00000024724
|
Olr852
|
olfactory receptor 852 |
| chr7_-_26983172 | 0.90 |
ENSRNOT00000089545
|
Txnrd1
|
thioredoxin reductase 1 |
| chr11_+_43687107 | 0.82 |
ENSRNOT00000048343
|
Olr1558
|
olfactory receptor 1558 |
| chrM_+_8599 | 0.81 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr17_+_47662532 | 0.75 |
ENSRNOT00000081314
|
Stard3nl
|
STARD3 N-terminal like |
| chr8_-_6034142 | 0.70 |
ENSRNOT00000014560
|
Birc2
|
baculoviral IAP repeat-containing 2 |
| chr17_-_89923423 | 0.64 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr2_-_210991509 | 0.62 |
ENSRNOT00000087329
|
Cyb561d1
|
cytochrome b561 family, member D1 |
| chr9_+_64095978 | 0.56 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr4_-_167089055 | 0.55 |
ENSRNOT00000050409
|
Tas2r113
|
taste receptor, type 2, member 113 |
| chr15_+_30525754 | 0.49 |
ENSRNOT00000086842
|
AABR07017745.2
|
|
| chr1_+_168204985 | 0.48 |
ENSRNOT00000049036
|
Olr75
|
olfactory receptor 75 |
| chr8_-_18408179 | 0.47 |
ENSRNOT00000040032
|
AABR07069371.1
|
|
| chr18_+_17550350 | 0.40 |
ENSRNOT00000078870
|
RGD1562608
|
similar to KIAA1328 protein |
| chr1_+_168092141 | 0.33 |
ENSRNOT00000072815
|
Or51t1
|
olfactory receptor, family 51, subfamily T, member 1 |
| chr18_-_53181503 | 0.33 |
ENSRNOT00000066548
|
Fbn2
|
fibrillin 2 |
| chr7_-_69982592 | 0.25 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr5_-_33892462 | 0.18 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chr1_-_47213749 | 0.14 |
ENSRNOT00000024656
|
Dynlt1
|
dynein light chain Tctex-type 1 |
| chr9_+_111028824 | 0.12 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_+_49578633 | 0.05 |
ENSRNOT00000072683
|
Crybb2
|
crystallin, beta B2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.4 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 5.6 | 16.9 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 5.0 | 15.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 4.6 | 13.9 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 3.5 | 10.5 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 3.5 | 10.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 3.4 | 13.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.9 | 11.8 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.9 | 11.7 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 2.6 | 7.9 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 2.3 | 6.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 2.2 | 17.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 1.9 | 5.7 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 1.7 | 5.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.6 | 16.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 1.5 | 23.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 1.2 | 3.6 | GO:0045575 | basophil activation(GO:0045575) |
| 1.2 | 3.5 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.1 | 10.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 1.1 | 12.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.1 | 3.2 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.0 | 3.9 | GO:0009597 | detection of virus(GO:0009597) |
| 0.9 | 8.3 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.8 | 2.4 | GO:1901581 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.8 | 15.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.8 | 15.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.8 | 11.4 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.7 | 7.5 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.7 | 2.2 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.5 | 1.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.5 | 7.1 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.4 | 9.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 9.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.4 | 2.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.4 | 10.5 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.4 | 6.3 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.4 | 1.5 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.4 | 6.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.3 | 8.9 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.3 | 11.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.3 | 5.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 1.0 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.3 | 2.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 6.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.2 | 2.4 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 9.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 0.7 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 0.6 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.2 | 1.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 8.2 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 7.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 7.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.9 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 2.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 8.5 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.1 | 2.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 11.1 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 0.3 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.1 | 2.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 13.7 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 1.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 11.1 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 10.0 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 2.9 | 11.8 | GO:0044393 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 2.6 | 10.4 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 2.2 | 6.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.9 | 7.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.6 | 8.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.4 | 6.9 | GO:0048476 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 1.3 | 7.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.8 | 5.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.8 | 2.4 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.7 | 27.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.7 | 16.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 2.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.3 | 15.8 | GO:0000791 | euchromatin(GO:0000791) |
| 0.2 | 14.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 16.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 15.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 3.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 9.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 7.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 10.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 4.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 6.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 15.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 19.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 9.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 31.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 8.3 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 1.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.1 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 32.0 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 2.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 3.9 | 19.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 3.4 | 13.7 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 3.4 | 16.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 3.4 | 10.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 2.8 | 13.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 2.8 | 11.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 2.7 | 13.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 2.1 | 10.5 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 2.1 | 14.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 1.9 | 7.5 | GO:0042806 | fucose binding(GO:0042806) |
| 1.7 | 10.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.5 | 15.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 1.1 | 6.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 1.0 | 9.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.0 | 6.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.8 | 2.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.8 | 9.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.7 | 11.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.7 | 3.5 | GO:1990715 | mRNA CDS binding(GO:1990715) C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 15.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.5 | 7.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 2.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 23.4 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.3 | 2.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 0.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 5.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 10.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 10.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 5.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 14.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 3.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 2.5 | GO:0016893 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters(GO:0016893) |
| 0.1 | 11.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 9.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 34.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 12.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 8.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 25.0 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 6.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 4.6 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 2.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 15.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.6 | 12.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.6 | 17.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.6 | 17.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.5 | 36.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.5 | 13.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 5.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 22.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 11.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.3 | 10.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 16.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 10.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 15.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 6.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 8.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 19.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 1.3 | 17.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.9 | 12.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.8 | 10.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.7 | 13.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 10.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.4 | 11.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 11.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 6.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 15.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.4 | 45.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.4 | 17.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 13.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 3.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 10.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 14.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 5.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 7.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 9.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 8.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 5.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 7.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 3.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 8.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 4.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |