Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
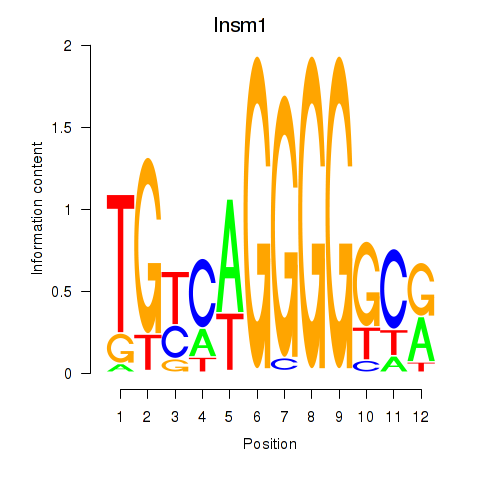
Results for Insm1
Z-value: 0.49
Transcription factors associated with Insm1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Insm1 motif
Sorted Z-values of Insm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_90999506 | 18.14 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr6_-_138249382 | 16.19 |
ENSRNOT00000085678
ENSRNOT00000006912 |
Ighm
|
immunoglobulin heavy constant mu |
| chr2_+_262914327 | 14.75 |
ENSRNOT00000029312
|
Negr1
|
neuronal growth regulator 1 |
| chr18_+_83777665 | 11.67 |
ENSRNOT00000018682
|
Cbln2
|
cerebellin 2 precursor |
| chr3_-_172537877 | 7.74 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr6_+_139405966 | 7.35 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr3_+_113251778 | 7.34 |
ENSRNOT00000083005
|
Map1a
|
microtubule-associated protein 1A |
| chr13_-_48284408 | 7.22 |
ENSRNOT00000085967
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_+_140602542 | 6.84 |
ENSRNOT00000085570
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr8_-_58647933 | 6.68 |
ENSRNOT00000038830
|
Tnfaip8l3
|
TNF alpha induced protein 8 like 3 |
| chr1_-_89360733 | 6.21 |
ENSRNOT00000028544
|
Mag
|
myelin-associated glycoprotein |
| chr5_+_142875773 | 5.71 |
ENSRNOT00000082120
ENSRNOT00000056496 |
Epha10
|
EPH receptor A10 |
| chr10_-_58693754 | 5.66 |
ENSRNOT00000071764
|
Pitpnm3
|
PITPNM family member 3 |
| chr6_+_57516713 | 5.51 |
ENSRNOT00000063874
|
Dgkb
|
diacylglycerol kinase, beta |
| chr19_-_19509087 | 4.84 |
ENSRNOT00000019301
|
Nkd1
|
naked cuticle homolog 1 |
| chr7_+_37812831 | 4.53 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr15_+_39644851 | 4.50 |
ENSRNOT00000081373
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr6_+_26537707 | 4.36 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chr3_+_154786215 | 4.34 |
ENSRNOT00000019787
|
Lbp
|
lipopolysaccharide binding protein |
| chr2_+_239415046 | 4.32 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr3_-_120087136 | 4.27 |
ENSRNOT00000078994
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr2_-_220101791 | 4.05 |
ENSRNOT00000022905
|
Plppr5
|
phospholipid phosphatase related 5 |
| chr19_+_55094585 | 4.01 |
ENSRNOT00000068452
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr6_-_92527711 | 3.90 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr1_-_197770669 | 3.78 |
ENSRNOT00000023563
|
Lat
|
linker for activation of T cells |
| chr20_+_2194709 | 3.68 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr17_-_90756048 | 3.63 |
ENSRNOT00000085669
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr6_+_147876237 | 3.45 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr12_+_22026075 | 3.14 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr19_+_54766589 | 2.87 |
ENSRNOT00000025894
|
Banp
|
Btg3 associated nuclear protein |
| chrX_+_14498119 | 2.86 |
ENSRNOT00000051135
|
Xk
|
X-linked Kx blood group |
| chr2_-_211322719 | 2.74 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr3_+_103753238 | 2.29 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr7_-_59514939 | 2.19 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr15_+_46784963 | 2.19 |
ENSRNOT00000015540
|
Xkr6
|
XK related 6 |
| chr4_-_153465203 | 2.14 |
ENSRNOT00000016776
|
Bid
|
BH3 interacting domain death agonist |
| chr2_+_251817694 | 2.08 |
ENSRNOT00000019964
|
RGD1560065
|
similar to RIKEN cDNA 2410004B18 |
| chr10_+_43538160 | 1.88 |
ENSRNOT00000089217
|
Cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr1_+_192025357 | 1.82 |
ENSRNOT00000025072
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr7_+_127081978 | 1.68 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr6_+_147876557 | 1.67 |
ENSRNOT00000080090
|
Tmem196
|
transmembrane protein 196 |
| chr20_+_46428124 | 1.65 |
ENSRNOT00000000327
|
Foxo3
|
forkhead box O3 |
| chr1_+_171820423 | 1.61 |
ENSRNOT00000047831
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr8_-_32328819 | 1.58 |
ENSRNOT00000074457
ENSRNOT00000070818 |
Aplp2
|
amyloid beta precursor like protein 2 |
| chrX_+_20216587 | 1.51 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr1_+_89202527 | 1.41 |
ENSRNOT00000028526
|
Sbsn
|
suprabasin |
| chr1_+_192025710 | 1.36 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr2_+_193286721 | 1.32 |
ENSRNOT00000074852
|
AABR07012323.1
|
|
| chrX_-_154722220 | 1.14 |
ENSRNOT00000089743
ENSRNOT00000087893 |
Fmr1
|
fragile X mental retardation 1 |
| chr4_+_113935492 | 1.13 |
ENSRNOT00000035329
|
LOC103692170
|
coiled-coil domain-containing protein 142 |
| chr3_-_148313810 | 1.06 |
ENSRNOT00000010762
|
Bcl2l1
|
Bcl2-like 1 |
| chr18_-_17396805 | 1.05 |
ENSRNOT00000082963
|
Tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr1_-_80630038 | 0.92 |
ENSRNOT00000025281
|
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr7_-_12424367 | 0.92 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr6_+_28323647 | 0.90 |
ENSRNOT00000089093
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr1_-_154170409 | 0.82 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr1_+_165382690 | 0.81 |
ENSRNOT00000023802
|
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr1_-_89488223 | 0.71 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_4293352 | 0.62 |
ENSRNOT00000047489
|
Olr984
|
olfactory receptor 984 |
| chr15_-_56970365 | 0.46 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr11_+_86890585 | 0.40 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr2_-_128002005 | 0.24 |
ENSRNOT00000018796
|
Pgrmc2
|
progesterone receptor membrane component 2 |
| chr18_-_61788859 | 0.21 |
ENSRNOT00000034075
ENSRNOT00000034069 |
Ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr2_+_189106039 | 0.05 |
ENSRNOT00000028210
|
Ube2q1
|
ubiquitin conjugating enzyme E2 Q1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Insm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 18.1 | GO:1904714 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 2.4 | 7.2 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 2.3 | 6.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.6 | 4.8 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.4 | 4.3 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 1.3 | 4.0 | GO:0071733 | mitral valve formation(GO:0003192) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 1.1 | 7.7 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 1.0 | 4.8 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.8 | 2.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.7 | 3.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.6 | 6.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.5 | 6.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.5 | 2.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.4 | 3.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 1.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.3 | 1.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.3 | 0.8 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.2 | 2.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 7.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 0.9 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 0.7 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.2 | 1.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 6.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 0.9 | GO:0044027 | hypermethylation of CpG island(GO:0044027) C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 5.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 4.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 5.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 2.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 16.4 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 4.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 9.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 3.8 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.1 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 4.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 3.1 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 1.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 4.4 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 2.1 | 6.2 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.7 | 8.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.6 | 3.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 7.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.2 | 4.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 3.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 13.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 7.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 6.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 4.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 1.1 | 6.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.0 | 5.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.9 | 4.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 2.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 6.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.4 | 5.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 4.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 3.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 7.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 1.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 18.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.1 | GO:0035197 | G-quadruplex RNA binding(GO:0002151) poly(G) binding(GO:0034046) siRNA binding(GO:0035197) |
| 0.1 | 3.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 7.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 6.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.9 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 4.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 7.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 9.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 9.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 3.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 6.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 16.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 6.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 7.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 5.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 6.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 4.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.9 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |