Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
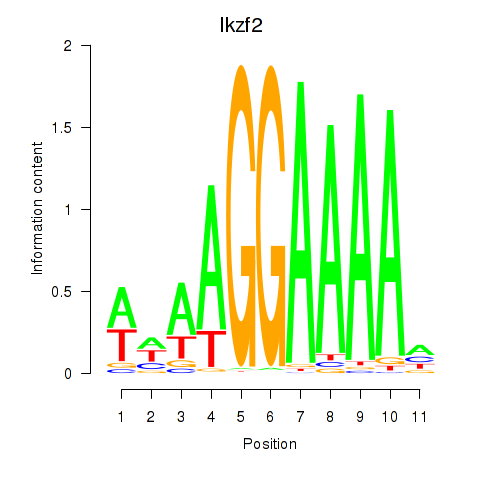
Results for Ikzf2
Z-value: 0.89
Transcription factors associated with Ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ikzf2
|
ENSRNOG00000027430 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ikzf2 | rn6_v1_chr9_-_76768770_76768806 | 0.61 | 1.7e-33 | Click! |
Activity profile of Ikzf2 motif
Sorted Z-values of Ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_19918644 | 24.46 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr6_+_99356509 | 24.06 |
ENSRNOT00000008416
|
Akap5
|
A-kinase anchoring protein 5 |
| chr17_-_43798383 | 22.55 |
ENSRNOT00000075069
|
LOC684828
|
similar to Histone H1.2 (H1 VAR.1) (H1c) |
| chr1_+_78711077 | 21.42 |
ENSRNOT00000064332
|
Slc1a5
|
solute carrier family 1 member 5 |
| chr9_-_76768770 | 20.87 |
ENSRNOT00000087779
ENSRNOT00000057849 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr6_-_131914028 | 19.33 |
ENSRNOT00000007602
|
Bcl11b
|
B-cell CLL/lymphoma 11B |
| chr4_+_147333056 | 18.18 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr7_-_107634287 | 16.26 |
ENSRNOT00000093672
ENSRNOT00000087116 |
Sla
|
src-like adaptor |
| chr2_+_236233239 | 15.81 |
ENSRNOT00000013694
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr9_+_52023295 | 15.62 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr8_-_107952530 | 14.93 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr2_+_150756185 | 14.67 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chrX_+_78259409 | 13.57 |
ENSRNOT00000049779
|
RGD1560455
|
similar to RIKEN cDNA A630033H20 gene |
| chr10_+_23914894 | 13.41 |
ENSRNOT00000071435
|
Ebf1
|
early B-cell factor 1 |
| chr15_-_33250546 | 12.95 |
ENSRNOT00000017857
|
RGD1565222
|
similar to RIKEN cDNA 4931414P19 |
| chrX_+_78042859 | 12.05 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr3_+_103192683 | 11.92 |
ENSRNOT00000037551
|
LOC100363452
|
ribosomal protein S8-like |
| chr1_+_141767940 | 11.80 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr13_-_89874008 | 11.43 |
ENSRNOT00000051368
|
Ptma
|
prothymosin alpha |
| chr20_+_27975549 | 11.31 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chrX_+_15155230 | 11.29 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr11_+_84745904 | 11.28 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr3_+_149790856 | 10.72 |
ENSRNOT00000075275
|
LOC100911637
|
high mobility group protein B1-like |
| chr1_-_260254600 | 10.65 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr18_+_30562178 | 10.21 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr10_+_23661343 | 10.06 |
ENSRNOT00000047970
|
Ebf1
|
early B-cell factor 1 |
| chr7_-_104541392 | 9.93 |
ENSRNOT00000078116
|
Fam49b
|
family with sequence similarity 49, member B |
| chr9_+_40975836 | 9.86 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr13_+_90244681 | 9.47 |
ENSRNOT00000078162
|
Cd84
|
CD84 molecule |
| chr4_-_29778039 | 9.37 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr9_+_71915421 | 9.35 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr14_+_43694183 | 9.31 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr12_-_51702730 | 9.24 |
ENSRNOT00000046920
|
Ttc28
|
tetratricopeptide repeat domain 28 |
| chr1_+_266255797 | 9.21 |
ENSRNOT00000027047
|
Trim8
|
tripartite motif-containing 8 |
| chr6_-_42630983 | 9.07 |
ENSRNOT00000071977
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr14_-_43694584 | 9.02 |
ENSRNOT00000041866
|
AABR07072400.1
|
|
| chr8_-_49158971 | 8.96 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr10_+_84135116 | 8.74 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chrX_-_29648359 | 8.60 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr11_-_14304603 | 8.58 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr8_-_118378460 | 8.48 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr8_+_118378059 | 8.37 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr1_+_253221812 | 8.31 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr6_-_77421286 | 7.97 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr3_+_93920013 | 7.67 |
ENSRNOT00000083527
|
Lmo2
|
LIM domain only 2 |
| chr7_-_51515373 | 7.45 |
ENSRNOT00000080285
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr3_+_44025300 | 7.39 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chrX_-_31851715 | 6.70 |
ENSRNOT00000068601
|
Vegfd
|
vascular endothelial growth factor D |
| chr3_+_113918629 | 6.68 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr13_+_46169963 | 6.36 |
ENSRNOT00000005212
|
Thsd7b
|
thrombospondin type 1 domain containing 7B |
| chr18_+_30487264 | 6.28 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr6_+_104475036 | 6.14 |
ENSRNOT00000070995
|
Susd6
|
sushi domain containing 6 |
| chr7_-_124020574 | 6.12 |
ENSRNOT00000055988
|
Poldip3
|
DNA polymerase delta interacting protein 3 |
| chr11_-_35749464 | 6.11 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr12_+_25450286 | 6.09 |
ENSRNOT00000092956
|
Gtf2i
|
general transcription factor II I |
| chr3_+_8873933 | 6.04 |
ENSRNOT00000030996
|
Nup188
|
nucleoporin 188 |
| chr3_+_93920447 | 5.97 |
ENSRNOT00000012625
|
Lmo2
|
LIM domain only 2 |
| chr18_+_30496318 | 5.75 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr14_+_88549947 | 5.74 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr10_-_92476109 | 5.71 |
ENSRNOT00000089029
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr17_+_10559680 | 5.53 |
ENSRNOT00000023320
|
Nop16
|
NOP16 nucleolar protein |
| chr6_+_28235695 | 5.50 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr7_-_120770435 | 5.47 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr4_+_100407658 | 5.46 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chr14_-_112946204 | 5.34 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr3_+_11554457 | 5.26 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr3_-_3855981 | 5.11 |
ENSRNOT00000079882
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr10_-_65066612 | 4.90 |
ENSRNOT00000043448
|
Myo18a
|
myosin XVIIIa |
| chr12_-_51965779 | 4.84 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr6_-_44361908 | 4.77 |
ENSRNOT00000009491
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr2_-_80667481 | 4.51 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr6_+_60566196 | 4.51 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chr2_-_235951275 | 4.43 |
ENSRNOT00000050291
|
AABR07013410.1
|
|
| chr2_-_208225888 | 4.39 |
ENSRNOT00000054860
|
AABR07012775.1
|
|
| chr8_+_52127632 | 4.19 |
ENSRNOT00000079797
|
Cadm1
|
cell adhesion molecule 1 |
| chr19_-_24614019 | 4.00 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr9_-_116222374 | 3.98 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr7_-_51515131 | 3.97 |
ENSRNOT00000006773
ENSRNOT00000041473 ENSRNOT00000050037 |
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr9_+_73433252 | 3.84 |
ENSRNOT00000092540
|
Map2
|
microtubule-associated protein 2 |
| chr6_-_26828972 | 3.81 |
ENSRNOT00000010932
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr4_+_66091641 | 3.78 |
ENSRNOT00000043147
|
AABR07060287.1
|
|
| chr3_+_61634005 | 3.72 |
ENSRNOT00000002145
|
Hoxd4
|
homeo box D4 |
| chr17_+_56109549 | 3.65 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr15_+_34493138 | 3.64 |
ENSRNOT00000089584
ENSRNOT00000027789 |
Nfatc4
|
nuclear factor of activated T-cells 4 |
| chr10_-_56409017 | 3.57 |
ENSRNOT00000020152
|
Fgf11
|
fibroblast growth factor 11 |
| chrX_+_84064427 | 3.51 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr7_-_125497691 | 3.40 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chr15_+_24479761 | 3.32 |
ENSRNOT00000016449
|
Ktn1
|
kinectin 1 |
| chr9_-_46206605 | 3.29 |
ENSRNOT00000018640
|
Tbc1d8
|
TBC1 domain family, member 8 |
| chrX_+_10430847 | 3.25 |
ENSRNOT00000047936
|
Rpl21
|
ribosomal protein L21 |
| chr18_-_53181503 | 3.23 |
ENSRNOT00000066548
|
Fbn2
|
fibrillin 2 |
| chr8_-_45375435 | 3.23 |
ENSRNOT00000010873
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr9_+_65172194 | 3.14 |
ENSRNOT00000040493
|
AC128084.1
|
|
| chr10_-_66848388 | 3.12 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr6_-_76552559 | 3.04 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr4_-_180722358 | 2.94 |
ENSRNOT00000040645
|
Itpr2
|
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr14_+_8080275 | 2.84 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr9_+_94316717 | 2.76 |
ENSRNOT00000077005
ENSRNOT00000092534 |
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr10_-_56365084 | 2.73 |
ENSRNOT00000068013
|
Polr2a
|
RNA polymerase II subunit A |
| chr13_+_57243877 | 2.60 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chrX_-_105417323 | 2.56 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr13_+_98311827 | 2.49 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr19_+_49495892 | 2.43 |
ENSRNOT00000015180
|
Atmin
|
ATM interactor |
| chr15_-_29465807 | 2.35 |
ENSRNOT00000075046
|
AABR07017635.1
|
|
| chrX_-_25590048 | 2.35 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr2_-_198120041 | 2.18 |
ENSRNOT00000079621
ENSRNOT00000028747 |
Plekho1
|
pleckstrin homology domain containing O1 |
| chr10_-_21265026 | 2.11 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr2_+_60920257 | 2.04 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr16_+_24980723 | 1.98 |
ENSRNOT00000082142
|
Tma16
|
translation machinery associated 16 homolog |
| chr2_+_31378743 | 1.84 |
ENSRNOT00000050384
|
AABR07007853.1
|
|
| chr10_+_17327275 | 1.78 |
ENSRNOT00000005486
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr5_-_154393697 | 1.68 |
ENSRNOT00000090019
|
Rpl11
|
ribosomal protein L11 |
| chr1_-_70485888 | 1.63 |
ENSRNOT00000020514
|
Olr7
|
olfactory receptor 7 |
| chr2_-_41784929 | 1.43 |
ENSRNOT00000086851
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr6_-_86713370 | 1.40 |
ENSRNOT00000005821
|
Klhl28
|
kelch-like family member 28 |
| chr7_+_122160171 | 1.31 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr2_+_198755262 | 0.92 |
ENSRNOT00000028807
|
Rbm8a
|
RNA binding motif protein 8A |
| chr4_+_28989115 | 0.38 |
ENSRNOT00000075326
|
Gng11
|
G protein subunit gamma 11 |
| chr3_-_59688692 | 0.32 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr1_+_172242504 | 0.29 |
ENSRNOT00000043135
|
Olr245
|
olfactory receptor 245 |
| chr4_-_34194764 | 0.24 |
ENSRNOT00000045270
|
Col28a1
|
collagen type XXVIII alpha 1 chain |
| chr9_-_94495333 | 0.24 |
ENSRNOT00000021507
|
Kcnj13
|
potassium voltage-gated channel subfamily J member 13 |
| chr14_+_8080565 | 0.00 |
ENSRNOT00000092395
|
Mapk10
|
mitogen activated protein kinase 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 6.0 | 24.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 5.9 | 35.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 5.0 | 14.9 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 3.9 | 15.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 3.3 | 9.9 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 3.1 | 21.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 3.0 | 9.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 2.8 | 22.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 2.8 | 8.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 2.4 | 9.5 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 1.9 | 11.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 1.7 | 8.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 4.9 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 1.6 | 4.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.5 | 6.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 1.5 | 4.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.3 | 8.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.3 | 24.5 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 1.2 | 3.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.0 | 11.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 1.0 | 6.1 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 1.0 | 13.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 1.0 | 6.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.0 | 5.7 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.9 | 5.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) C-5 methylation of cytosine(GO:0090116) |
| 0.9 | 6.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.8 | 4.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.8 | 3.2 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.8 | 25.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.8 | 11.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.7 | 5.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 | 11.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.5 | 14.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.5 | 9.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.4 | 6.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 2.9 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.4 | 4.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 2.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 6.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.3 | 9.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.3 | 5.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 | 5.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.3 | 1.7 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.3 | 2.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 4.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.3 | 3.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.3 | 2.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 8.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 5.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 5.7 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 2.0 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 11.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.3 | 9.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 8.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.2 | 2.7 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.2 | 2.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 16.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 2.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 2.5 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 22.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 3.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.1 | 3.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 9.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 5.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 2.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 6.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 3.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 4.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 3.7 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 7.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 7.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 2.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 7.7 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 1.4 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 2.8 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 6.1 | GO:0006974 | cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 | 4.3 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 2.0 | 6.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.9 | 5.7 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 1.8 | 9.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 1.6 | 24.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.6 | 9.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.3 | 15.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.9 | 11.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.8 | 25.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.8 | 5.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.7 | 9.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.6 | 8.3 | GO:0097431 | mitotic spindle pole(GO:0097431) mitotic spindle midzone(GO:1990023) |
| 0.5 | 14.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.5 | 5.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.4 | 2.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 6.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.3 | 3.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 2.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 14.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 4.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 24.5 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 2.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 11.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 18.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 16.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 11.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 4.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 11.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 7.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 25.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 4.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 5.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 48.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 8.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 11.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 6.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 9.9 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 19.0 | GO:0097458 | neuron part(GO:0097458) |
| 0.0 | 8.1 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 7.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 4.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 3.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 20.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.6 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 3.3 | 9.9 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 3.0 | 12.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 2.6 | 18.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 2.2 | 24.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 1.9 | 39.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 1.8 | 9.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.8 | 21.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.6 | 9.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 1.3 | 10.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 1.3 | 3.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.2 | 9.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 1.1 | 15.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 5.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 1.0 | 9.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.8 | 5.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.7 | 6.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.7 | 2.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.7 | 7.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.7 | 8.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.6 | 5.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.5 | 8.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 5.7 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 11.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.5 | 11.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 13.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.3 | 2.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 2.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 6.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 2.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 8.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 16.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 5.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 22.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 11.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 6.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 3.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 5.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 2.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 4.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 27.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 4.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 5.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 52.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 15.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 4.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 6.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 25.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 16.8 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 6.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 3.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 27.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.4 | 21.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.4 | 15.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.4 | 10.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.4 | 17.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 4.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 20.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 20.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 3.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 11.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 13.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 8.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 6.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 4.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 5.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 5.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 3.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 7.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 12.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.8 | 9.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.7 | 24.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.7 | 11.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 21.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 10.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.5 | 14.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.5 | 11.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 39.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.4 | 5.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.4 | 15.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 9.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 4.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 4.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 2.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 6.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 3.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 2.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 2.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 4.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 7.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 6.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 4.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 3.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |