Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
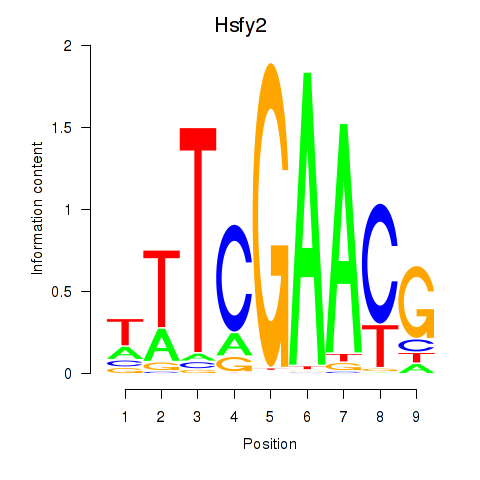
Results for Hsfy2
Z-value: 0.77
Transcription factors associated with Hsfy2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsfy2
|
ENSRNOG00000038569 | heat shock transcription factor, Y linked 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsfy2 | rn6_v1_chr9_-_63291350_63291350 | -0.26 | 1.8e-06 | Click! |
Activity profile of Hsfy2 motif
Sorted Z-values of Hsfy2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_24456976 | 42.76 |
ENSRNOT00000004900
|
Ucp1
|
uncoupling protein 1 |
| chr3_-_91217491 | 18.16 |
ENSRNOT00000006115
|
Rag1
|
recombination activating 1 |
| chr13_-_91981432 | 14.97 |
ENSRNOT00000004637
|
AABR07021804.1
|
|
| chr2_+_200785507 | 11.95 |
ENSRNOT00000046942
|
Hao2
|
hydroxyacid oxidase 2 |
| chr16_-_74122889 | 11.75 |
ENSRNOT00000025763
|
Plat
|
plasminogen activator, tissue type |
| chr8_+_49282460 | 11.52 |
ENSRNOT00000021488
|
Cd3d
|
CD3d molecule |
| chr2_+_60337667 | 11.29 |
ENSRNOT00000024035
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr4_-_145390447 | 11.14 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr5_-_159962676 | 10.78 |
ENSRNOT00000013550
|
Clcnkb
|
chloride voltage-gated channel Kb |
| chr17_-_9762813 | 10.76 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr20_-_31598118 | 10.53 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr10_-_47059216 | 10.08 |
ENSRNOT00000007092
|
Shmt1
|
serine hydroxymethyltransferase 1 |
| chr20_-_31597830 | 10.00 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chrX_-_1848904 | 9.76 |
ENSRNOT00000010984
|
Rgn
|
regucalcin |
| chr4_+_71621729 | 9.49 |
ENSRNOT00000022275
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr1_+_150797084 | 9.38 |
ENSRNOT00000018990
|
Nox4
|
NADPH oxidase 4 |
| chr9_-_15410943 | 9.15 |
ENSRNOT00000074217
|
Ccnd3
|
cyclin D3 |
| chr5_-_159962218 | 8.78 |
ENSRNOT00000050729
|
Clcnkb
|
chloride voltage-gated channel Kb |
| chr2_-_235161263 | 8.68 |
ENSRNOT00000080235
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr18_+_38292701 | 8.36 |
ENSRNOT00000037796
|
Scgb3a2
|
secretoglobin, family 3A, member 2 |
| chr9_-_20154077 | 8.12 |
ENSRNOT00000082904
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr4_-_176679815 | 8.07 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr8_-_107952530 | 7.79 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr9_+_61692154 | 7.58 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr14_+_91783514 | 7.50 |
ENSRNOT00000080753
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr1_-_22625204 | 7.33 |
ENSRNOT00000021694
|
Vnn1
|
vanin 1 |
| chr1_-_47307488 | 7.33 |
ENSRNOT00000090033
|
Ezr
|
ezrin |
| chr1_+_141821916 | 7.15 |
ENSRNOT00000071574
|
Echs1
|
enoyl-CoA hydratase, short chain 1 |
| chr13_-_104080631 | 7.05 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr19_+_55982740 | 6.91 |
ENSRNOT00000021397
|
Dpep1
|
dipeptidase 1 (renal) |
| chr10_-_31493419 | 6.89 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr10_+_71202456 | 6.88 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr16_-_49003246 | 6.82 |
ENSRNOT00000089501
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_+_145594575 | 6.79 |
ENSRNOT00000029819
|
Irak2
|
interleukin-1 receptor-associated kinase 2 |
| chr7_-_143793774 | 6.76 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr9_+_112293388 | 6.74 |
ENSRNOT00000020767
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr4_+_70828894 | 6.62 |
ENSRNOT00000064892
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr12_+_40466495 | 6.56 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr1_+_189364288 | 6.51 |
ENSRNOT00000080338
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr20_-_4070721 | 6.40 |
ENSRNOT00000000523
|
RT1-Ba
|
RT1 class II, locus Ba |
| chr10_-_15235740 | 6.28 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr1_-_216663720 | 6.06 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chrX_+_156319687 | 5.76 |
ENSRNOT00000091147
ENSRNOT00000079378 |
Fam3a
|
family with sequence similarity 3, member A |
| chr9_+_16924520 | 5.75 |
ENSRNOT00000025094
|
Slc22a7
|
solute carrier family 22 member 7 |
| chr6_+_107531528 | 5.70 |
ENSRNOT00000077555
|
Acot3
|
acyl-CoA thioesterase 3 |
| chr4_-_115015965 | 5.68 |
ENSRNOT00000014603
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr16_+_54319377 | 5.67 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr1_+_125229469 | 5.65 |
ENSRNOT00000021884
|
Mcee
|
methylmalonyl CoA epimerase |
| chr1_+_199682688 | 5.65 |
ENSRNOT00000027265
ENSRNOT00000080872 |
Slc5a2
|
solute carrier family 5 member 2 |
| chr10_-_94576512 | 5.57 |
ENSRNOT00000035474
|
Icam2
|
intercellular adhesion molecule 2 |
| chr3_+_129462738 | 5.52 |
ENSRNOT00000077755
|
AABR07053830.1
|
|
| chr10_-_15465404 | 5.42 |
ENSRNOT00000077826
ENSRNOT00000027593 |
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr1_+_191829555 | 5.30 |
ENSRNOT00000067138
|
Scnn1b
|
sodium channel epithelial 1 beta subunit |
| chr7_-_143793970 | 5.29 |
ENSRNOT00000016205
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr2_+_26240385 | 5.28 |
ENSRNOT00000024292
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr5_+_34007920 | 5.24 |
ENSRNOT00000009611
|
Ttpa
|
alpha tocopherol transfer protein |
| chr14_+_85113578 | 5.24 |
ENSRNOT00000011158
|
Nipsnap1
|
nipsnap homolog 1 |
| chr19_-_11302938 | 5.20 |
ENSRNOT00000038212
|
AC128848.1
|
|
| chr10_+_86399827 | 5.18 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr5_+_4982348 | 5.17 |
ENSRNOT00000010369
|
Lactb2
|
lactamase, beta 2 |
| chr1_+_104576589 | 5.06 |
ENSRNOT00000046529
|
Nav2
|
neuron navigator 2 |
| chr7_-_130405347 | 4.99 |
ENSRNOT00000013985
|
Cpt1b
|
carnitine palmitoyltransferase 1B |
| chr2_-_23256158 | 4.96 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr1_-_226935689 | 4.95 |
ENSRNOT00000038807
|
Tmem109
|
transmembrane protein 109 |
| chr15_+_32811135 | 4.85 |
ENSRNOT00000067689
|
AABR07017902.1
|
|
| chr3_-_67668772 | 4.77 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr16_+_54291251 | 4.72 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr4_+_71650575 | 4.62 |
ENSRNOT00000033775
|
Tmem139
|
transmembrane protein 139 |
| chr12_+_52452273 | 4.61 |
ENSRNOT00000056680
ENSRNOT00000088381 |
Pxmp2
|
peroxisomal membrane protein 2 |
| chr16_+_50022998 | 4.57 |
ENSRNOT00000087986
|
Tlr3
|
toll-like receptor 3 |
| chr20_+_13760810 | 4.51 |
ENSRNOT00000081140
ENSRNOT00000080203 |
Gstt2
|
glutathione S-transferase, theta 2 |
| chr3_+_72329967 | 4.50 |
ENSRNOT00000090256
|
Slc43a3
|
solute carrier family 43, member 3 |
| chr18_-_51651267 | 4.48 |
ENSRNOT00000020325
|
Aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr8_-_52937972 | 4.42 |
ENSRNOT00000007789
|
Nnmt
|
nicotinamide N-methyltransferase |
| chr11_+_47243342 | 4.42 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chrX_-_13601069 | 4.40 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr14_+_71542057 | 4.34 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr5_+_126670825 | 4.32 |
ENSRNOT00000012201
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr3_-_79728879 | 4.29 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr17_-_43689311 | 4.16 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr13_-_70573572 | 4.10 |
ENSRNOT00000092339
|
Lamc2
|
laminin subunit gamma 2 |
| chr1_-_61872975 | 4.00 |
ENSRNOT00000078809
|
AABR07001910.1
|
|
| chr1_-_207811008 | 3.92 |
ENSRNOT00000080506
|
Clrn3
|
clarin 3 |
| chr8_-_62424303 | 3.91 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr4_+_96562725 | 3.90 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr19_-_57614558 | 3.90 |
ENSRNOT00000025948
|
RGD1562218
|
similar to RIKEN cDNA 0610039J04 |
| chr7_-_3386522 | 3.88 |
ENSRNOT00000010760
|
Mettl7b
|
methyltransferase like 7B |
| chr5_+_141491223 | 3.88 |
ENSRNOT00000034839
|
Rhbdl2
|
rhomboid like 2 |
| chr1_-_226887156 | 3.84 |
ENSRNOT00000054809
ENSRNOT00000028347 |
Cd6
|
Cd6 molecule |
| chr2_-_47096961 | 3.83 |
ENSRNOT00000077401
|
Itga2
|
integrin alpha 2 |
| chr10_-_70337532 | 3.82 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr7_-_11319516 | 3.77 |
ENSRNOT00000027784
|
Apba3
|
amyloid beta precursor protein binding family A member 3 |
| chr2_+_84275884 | 3.75 |
ENSRNOT00000014439
|
Dap
|
death-associated protein |
| chr2_-_117666683 | 3.70 |
ENSRNOT00000015479
|
AABR07009931.1
|
|
| chr5_-_172307431 | 3.67 |
ENSRNOT00000018453
|
Fam213b
|
family with sequence similarity 213, member B |
| chr10_+_35133252 | 3.65 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr7_-_63578490 | 3.65 |
ENSRNOT00000007295
|
Rassf3
|
Ras association domain family member 3 |
| chr2_+_128461224 | 3.65 |
ENSRNOT00000018872
|
Jade1
|
jade family PHD finger 1 |
| chr4_+_109477920 | 3.58 |
ENSRNOT00000008468
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr1_+_228142778 | 3.57 |
ENSRNOT00000028517
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr3_-_150108898 | 3.51 |
ENSRNOT00000022914
|
Pxmp4
|
peroxisomal membrane protein 4 |
| chr4_+_153874852 | 3.50 |
ENSRNOT00000079744
|
Slc6a13
|
solute carrier family 6 member 13 |
| chrM_+_7006 | 3.46 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr4_+_120672152 | 3.45 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr6_+_43884678 | 3.43 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr10_-_78156477 | 3.36 |
ENSRNOT00000083009
|
Stxbp4
|
syntaxin binding protein 4 |
| chr20_+_48504264 | 3.35 |
ENSRNOT00000087740
|
Cdc40
|
cell division cycle 40 |
| chr1_-_76780230 | 3.32 |
ENSRNOT00000002046
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr16_-_68968248 | 3.31 |
ENSRNOT00000016885
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr5_-_4975436 | 3.29 |
ENSRNOT00000062006
|
Xkr9
|
XK related 9 |
| chr1_+_83653234 | 3.18 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr17_+_60059949 | 3.14 |
ENSRNOT00000025458
|
Mpp7
|
membrane palmitoylated protein 7 |
| chr4_+_155321553 | 3.08 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr1_-_85317968 | 3.04 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chrX_-_74968405 | 3.04 |
ENSRNOT00000035653
|
RGD1561931
|
similar to KIAA2022 protein |
| chr6_+_99402360 | 3.02 |
ENSRNOT00000078498
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr6_-_26486695 | 3.01 |
ENSRNOT00000073236
|
Krtcap3
|
keratinocyte associated protein 3 |
| chr10_-_76039964 | 3.00 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr9_+_9988055 | 2.93 |
ENSRNOT00000086575
ENSRNOT00000073000 |
Slc25a23
|
solute carrier family 25 member 23 |
| chrX_+_1787266 | 2.91 |
ENSRNOT00000011183
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr2_+_127845034 | 2.90 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr2_-_192960294 | 2.90 |
ENSRNOT00000012420
|
LOC102552326
|
late cornified envelope protein 5A-like |
| chr15_+_56757315 | 2.88 |
ENSRNOT00000078013
|
Esd
|
esterase D |
| chr1_-_263885169 | 2.87 |
ENSRNOT00000030782
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr17_+_72218769 | 2.87 |
ENSRNOT00000041346
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr3_+_175708519 | 2.87 |
ENSRNOT00000085983
|
AC115322.1
|
|
| chr9_-_49950093 | 2.86 |
ENSRNOT00000023014
|
Fhl2
|
four and a half LIM domains 2 |
| chr5_-_133786403 | 2.82 |
ENSRNOT00000081051
ENSRNOT00000010318 |
Cmpk1
|
cytidine/uridine monophosphate kinase 1 |
| chr12_-_22138382 | 2.81 |
ENSRNOT00000001899
|
Lrch4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr8_-_87158368 | 2.80 |
ENSRNOT00000077071
|
Col12a1
|
collagen type XII alpha 1 chain |
| chr8_-_28044876 | 2.77 |
ENSRNOT00000072152
|
Acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr15_+_32276947 | 2.76 |
ENSRNOT00000068094
|
AABR07017902.1
|
|
| chr14_+_13192347 | 2.75 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr16_-_49820235 | 2.74 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_-_77865870 | 2.74 |
ENSRNOT00000017353
|
Ehd2
|
EH-domain containing 2 |
| chr10_-_109747987 | 2.72 |
ENSRNOT00000054958
|
P4hb
|
prolyl 4-hydroxylase subunit beta |
| chrX_-_72077590 | 2.72 |
ENSRNOT00000004278
|
Rps4x
|
ribosomal protein S4, X-linked |
| chrX_-_128268285 | 2.71 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chrX_-_84768463 | 2.69 |
ENSRNOT00000088570
|
Chm
|
CHM, Rab escort protein 1 |
| chr17_+_90696019 | 2.67 |
ENSRNOT00000003438
|
Gpr137b
|
G protein-coupled receptor 137B |
| chr5_+_135997052 | 2.66 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr16_-_37177033 | 2.66 |
ENSRNOT00000014015
|
Fbxo8
|
F-box protein 8 |
| chr4_-_156427755 | 2.62 |
ENSRNOT00000085675
ENSRNOT00000014030 |
LOC103690024
|
peroxisomal targeting signal 1 receptor-like |
| chr4_-_157008947 | 2.59 |
ENSRNOT00000073341
|
Pex5
|
peroxisomal biogenesis factor 5 |
| chr10_+_88764732 | 2.57 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr2_+_104854572 | 2.56 |
ENSRNOT00000064828
ENSRNOT00000090912 |
Hltf
|
helicase-like transcription factor |
| chrX_+_140175861 | 2.55 |
ENSRNOT00000071222
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr14_+_2613406 | 2.51 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr6_+_64649194 | 2.50 |
ENSRNOT00000039776
|
AABR07064102.1
|
|
| chr19_+_10596960 | 2.49 |
ENSRNOT00000021769
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr9_-_10054359 | 2.48 |
ENSRNOT00000072001
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr20_+_3167079 | 2.46 |
ENSRNOT00000001035
|
RT1-N3
|
RT1 class Ib, locus N3 |
| chr10_+_40438356 | 2.45 |
ENSRNOT00000078910
|
Gm2a
|
GM2 ganglioside activator |
| chr1_+_166893734 | 2.44 |
ENSRNOT00000026702
|
Phox2a
|
paired-like homeobox 2a |
| chr8_-_23099042 | 2.41 |
ENSRNOT00000019115
|
Ecsit
|
ECSIT signalling integrator |
| chr1_+_259926537 | 2.41 |
ENSRNOT00000073537
|
NEWGENE_1306399
|
cyclin J |
| chr2_-_192671059 | 2.40 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr12_-_37480297 | 2.39 |
ENSRNOT00000001397
|
Tmed2
|
transmembrane p24 trafficking protein 2 |
| chr9_-_99651813 | 2.36 |
ENSRNOT00000022089
|
Ndufa10
|
NADH:ubiquinone oxidoreductase subunit A10 |
| chr1_+_36320461 | 2.34 |
ENSRNOT00000023659
|
Srd5a1
|
steroid 5 alpha-reductase 1 |
| chr18_+_32273770 | 2.32 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr7_-_126701872 | 2.31 |
ENSRNOT00000041057
|
Pkdrej
|
polycystin (PKD) family receptor for egg jelly |
| chr11_+_33863500 | 2.31 |
ENSRNOT00000072384
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chr9_+_95285592 | 2.29 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_-_53140637 | 2.28 |
ENSRNOT00000060492
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr11_-_89239798 | 2.27 |
ENSRNOT00000089517
|
Spidr
|
scaffolding protein involved in DNA repair |
| chr19_-_10620671 | 2.27 |
ENSRNOT00000021842
|
Ccl17
|
C-C motif chemokine ligand 17 |
| chr17_+_72160735 | 2.26 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_100755682 | 2.26 |
ENSRNOT00000035748
|
Vrk3
|
vaccinia related kinase 3 |
| chr10_-_87300728 | 2.25 |
ENSRNOT00000036023
ENSRNOT00000081709 |
Krt10
|
keratin 10 |
| chr8_-_55037604 | 2.25 |
ENSRNOT00000059169
|
Sdhd
|
succinate dehydrogenase complex subunit D |
| chr12_-_11215649 | 2.23 |
ENSRNOT00000042474
ENSRNOT00000076369 |
Cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr2_-_193136520 | 2.22 |
ENSRNOT00000042142
|
Kprp
|
keratinocyte proline-rich protein |
| chr1_+_57692836 | 2.20 |
ENSRNOT00000083968
ENSRNOT00000019358 |
Chd1
|
chromodomain helicase DNA binding protein 1 |
| chr14_+_84447885 | 2.20 |
ENSRNOT00000009150
|
Gatsl3
|
GATS protein-like 3 |
| chr15_+_105640097 | 2.19 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chrX_+_123770337 | 2.19 |
ENSRNOT00000092554
|
Nkap
|
NFKB activating protein |
| chr8_-_33463467 | 2.17 |
ENSRNOT00000041038
|
Kcnj5
|
potassium voltage-gated channel subfamily J member 5 |
| chr1_-_100669684 | 2.15 |
ENSRNOT00000091760
|
Myh14
|
myosin heavy chain 14 |
| chr9_-_61033553 | 2.15 |
ENSRNOT00000002864
ENSRNOT00000082681 |
Gtf3c3
|
general transcription factor IIIC subunit 3 |
| chr15_+_31446983 | 2.14 |
ENSRNOT00000089506
|
AABR07017825.6
|
|
| chr19_+_38768467 | 2.14 |
ENSRNOT00000027346
|
Cdh1
|
cadherin 1 |
| chr10_-_56267213 | 2.14 |
ENSRNOT00000017201
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr11_-_82938357 | 2.12 |
ENSRNOT00000035945
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_34546988 | 2.11 |
ENSRNOT00000072577
|
Adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr10_-_57275708 | 2.11 |
ENSRNOT00000005370
|
Pfn1
|
profilin 1 |
| chr10_+_10530365 | 2.10 |
ENSRNOT00000003843
|
Eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chrX_-_77061604 | 2.10 |
ENSRNOT00000083643
ENSRNOT00000089386 |
Magt1
|
magnesium transporter 1 |
| chr14_+_22251499 | 2.10 |
ENSRNOT00000087991
ENSRNOT00000002705 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr1_+_257076221 | 2.10 |
ENSRNOT00000044895
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr1_-_168471161 | 2.06 |
ENSRNOT00000055227
|
Olr94
|
olfactory receptor 94 |
| chr20_+_3875706 | 2.04 |
ENSRNOT00000036900
|
RT1-Ha
|
RT1 class II, locus Ha |
| chr19_-_44101365 | 2.02 |
ENSRNOT00000082182
|
Tmem170a
|
transmembrane protein 170A |
| chr2_+_44664124 | 2.00 |
ENSRNOT00000066098
|
Plpp1
|
phospholipid phosphatase 1 |
| chr8_+_132441285 | 1.99 |
ENSRNOT00000087488
ENSRNOT00000068233 |
Lars2
|
leucyl-tRNA synthetase 2 |
| chr10_+_96639924 | 1.99 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr17_+_35435079 | 1.98 |
ENSRNOT00000074800
|
Exoc2
|
exocyst complex component 2 |
| chr4_-_78074906 | 1.97 |
ENSRNOT00000080654
|
Zfp467
|
zinc finger protein 467 |
| chr1_+_219000844 | 1.97 |
ENSRNOT00000022486
|
Kmt5b
|
lysine methyltransferase 5B |
| chr7_-_117680004 | 1.96 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsfy2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 4.0 | 12.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 4.0 | 12.0 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 3.8 | 11.3 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) |
| 3.6 | 10.8 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 3.4 | 10.1 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 3.3 | 9.8 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 2.9 | 11.8 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 2.6 | 5.2 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 2.6 | 39.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 2.6 | 7.8 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 2.4 | 19.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 2.3 | 6.9 | GO:1901146 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.9 | 5.7 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 1.9 | 7.5 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 1.8 | 7.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.8 | 11.0 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 1.7 | 5.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 1.5 | 4.6 | GO:0006227 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 1.5 | 4.5 | GO:0009751 | response to salicylic acid(GO:0009751) |
| 1.3 | 5.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.1 | 4.6 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 1.1 | 4.3 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.0 | 6.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.0 | 3.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 1.0 | 6.8 | GO:0034201 | response to oleic acid(GO:0034201) |
| 1.0 | 2.9 | GO:1902741 | response to acetate(GO:0010034) type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 1.0 | 4.8 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.9 | 2.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.9 | 16.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.9 | 8.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.9 | 4.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.9 | 2.6 | GO:1901093 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.9 | 3.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.9 | 6.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.8 | 11.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.8 | 2.4 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.8 | 3.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.8 | 2.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.7 | 9.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.7 | 10.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.7 | 2.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.6 | 2.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.6 | 2.6 | GO:0060375 | development of secondary male sexual characteristics(GO:0046544) regulation of mast cell differentiation(GO:0060375) |
| 0.6 | 1.9 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.6 | 3.8 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.6 | 20.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.6 | 6.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.6 | 7.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.6 | 3.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.6 | 1.8 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.6 | 3.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.6 | 6.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.6 | 2.3 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.6 | 4.5 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.6 | 3.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.5 | 2.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.5 | 1.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.5 | 1.6 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.5 | 1.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.5 | 2.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 6.9 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.5 | 5.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.5 | 3.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 2.9 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.5 | 1.9 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.5 | 5.7 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.5 | 3.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.5 | 2.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 1.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.4 | 5.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.4 | 5.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.4 | 4.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.4 | 5.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.4 | 6.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 5.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.4 | 1.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.4 | 1.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.4 | 2.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 0.8 | GO:0061198 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.4 | 6.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.4 | 1.5 | GO:1900063 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) regulation of peroxisome organization(GO:1900063) |
| 0.4 | 1.1 | GO:0060846 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.4 | 2.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 3.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 10.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.3 | 0.7 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 3.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.3 | 4.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 2.3 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.3 | 3.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 1.6 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 2.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 4.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 1.2 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.3 | 1.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) threonine metabolic process(GO:0006566) |
| 0.3 | 1.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.3 | 2.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.3 | 2.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 2.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 1.4 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.3 | 2.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 2.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 5.7 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.3 | 1.9 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.3 | 2.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 0.5 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 | 0.8 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.3 | 3.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 1.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.2 | 1.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 2.7 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 1.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 1.0 | GO:0002337 | B-1 B cell differentiation(GO:0001923) B-1a B cell differentiation(GO:0002337) |
| 0.2 | 3.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 3.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 2.9 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.2 | 3.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 1.6 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 5.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.2 | 7.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 4.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 4.3 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.2 | 1.3 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.2 | 1.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 2.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 9.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 1.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 12.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 0.4 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.2 | 2.3 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 1.0 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 1.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 2.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.7 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 0.5 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 1.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 5.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.4 | GO:0061193 | taste bud development(GO:0061193) |
| 0.2 | 0.5 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 3.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 1.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 0.8 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 0.5 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.2 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 6.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 1.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 5.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 3.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 2.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 3.7 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.5 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 2.9 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 0.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 2.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 2.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 2.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 6.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 2.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.0 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 2.4 | GO:0060716 | vesicle targeting(GO:0006903) labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 1.8 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 1.1 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 3.8 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.4 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 2.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.3 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 2.7 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 2.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 3.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.6 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.1 | 0.8 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 5.7 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 3.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 3.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 3.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.0 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 2.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.2 | GO:1903960 | negative regulation of anion channel activity(GO:0010360) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.1 | 3.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 4.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 1.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 2.1 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 2.3 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 1.1 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 2.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.8 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.5 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.5 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.2 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 3.0 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.5 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.5 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 1.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.4 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 18.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.3 | GO:0036398 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 1.4 | 4.3 | GO:0071914 | prominosome(GO:0071914) |
| 1.4 | 11.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.3 | 3.8 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.3 | 3.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.0 | 4.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.9 | 9.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.9 | 3.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 3.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.8 | 2.3 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.7 | 2.8 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.7 | 2.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.7 | 2.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 3.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.5 | 2.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.5 | 8.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 7.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.5 | 5.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 2.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 5.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.4 | 26.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.4 | 2.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 6.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 1.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 1.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 2.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 20.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 3.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 7.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 1.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 2.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 2.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.3 | 5.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 17.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 4.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 2.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 5.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 5.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 11.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 10.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 3.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 21.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 8.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 58.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 10.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 8.4 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 1.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 0.7 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.2 | 0.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 2.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 13.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 41.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.7 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 6.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 2.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 1.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 2.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 2.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 4.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.4 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.2 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 6.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 2.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 4.9 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 4.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 5.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 12.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 5.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 32.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 1.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 4.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 5.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0031672 | A band(GO:0031672) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 42.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 4.0 | 12.0 | GO:0052852 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 3.4 | 10.1 | GO:0070905 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) serine binding(GO:0070905) |
| 2.3 | 6.9 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 2.3 | 11.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 2.2 | 10.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.7 | 6.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.5 | 4.6 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 1.5 | 4.4 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 1.4 | 5.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.4 | 5.4 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.3 | 6.6 | GO:0070404 | NADH binding(GO:0070404) |
| 1.2 | 5.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.2 | 8.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 1.1 | 4.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.0 | 5.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.0 | 9.4 | GO:0016175 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.0 | 19.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.0 | 2.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.9 | 7.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.8 | 6.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.8 | 3.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.8 | 2.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.7 | 2.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.7 | 2.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.7 | 2.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.6 | 5.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 5.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.6 | 13.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.6 | 2.3 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.6 | 3.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.6 | 3.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 5.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 2.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 2.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 9.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 1.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.5 | 6.7 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.5 | 1.5 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 2.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.5 | 1.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.5 | 2.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 6.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 5.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.4 | 10.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.4 | 1.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.4 | 1.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.4 | 1.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 4.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.4 | 1.4 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.3 | 3.4 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.3 | 3.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 1.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 2.9 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 2.0 | GO:0031727 | CCR1 chemokine receptor binding(GO:0031726) CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 5.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.3 | 4.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 12.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 7.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 8.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 3.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 3.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.3 | 0.8 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 10.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 1.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 2.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 6.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 5.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 5.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 2.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 3.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 3.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 1.6 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 3.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 5.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 2.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 5.2 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.2 | 2.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 4.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 21.8 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.2 | 1.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 4.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.2 | 2.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 8.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 1.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 24.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 2.1 | GO:0015095 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 3.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 1.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 3.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 3.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 8.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 5.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 8.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 5.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 2.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 4.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 6.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 10.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 2.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.5 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 2.4 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 3.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 7.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 6.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.5 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 3.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 6.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 13.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.7 | GO:0019840 | isoprenoid binding(GO:0019840) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 5.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 4.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.3 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 17.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.6 | 17.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 21.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.4 | 24.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.4 | 4.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 11.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.4 | 5.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 12.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 2.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 6.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 9.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 8.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 4.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 8.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 3.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 2.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 5.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 3.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 10.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 11.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.0 | 3.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.9 | 12.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.9 | 11.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.8 | 7.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 6.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.7 | 58.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.7 | 6.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.6 | 6.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.5 | 15.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.5 | 5.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 8.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 24.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 8.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 7.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.3 | 9.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 5.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 3.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 5.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 7.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 4.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 3.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 9.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 3.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 1.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 8.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 1.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.2 | 10.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 2.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 2.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 10.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 2.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 3.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 6.6 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 4.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 4.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 7.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 5.6 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.4 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.1 | 7.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 3.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 2.3 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |