Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
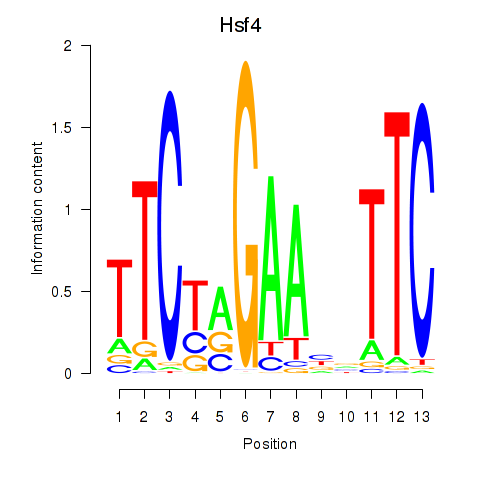
Results for Hsf4
Z-value: 0.94
Transcription factors associated with Hsf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf4
|
ENSRNOG00000015253 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf4 | rn6_v1_chr19_+_37226186_37226188 | -0.29 | 2.0e-07 | Click! |
Activity profile of Hsf4 motif
Sorted Z-values of Hsf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_170475831 | 39.31 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr17_-_44834305 | 30.10 |
ENSRNOT00000084303
|
Hist1h2bd
|
histone cluster 1, H2bd |
| chr9_-_14668297 | 30.01 |
ENSRNOT00000042404
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr8_-_49301125 | 29.19 |
ENSRNOT00000091190
|
Cd3e
|
CD3e molecule |
| chr11_+_86094567 | 24.46 |
ENSRNOT00000086514
|
LOC100361706
|
lambda-chain C1-region-like |
| chr14_+_3058993 | 24.13 |
ENSRNOT00000002807
|
Gfi1
|
growth factor independent 1 transcriptional repressor |
| chr7_-_116106368 | 23.43 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr8_+_132828091 | 20.89 |
ENSRNOT00000008269
|
Ccr9
|
C-C motif chemokine receptor 9 |
| chr1_-_89045586 | 20.80 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr14_-_82347679 | 20.70 |
ENSRNOT00000032972
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr3_+_170994038 | 19.84 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr18_+_25749098 | 19.79 |
ENSRNOT00000027771
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr4_+_170958196 | 18.96 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr13_-_84452181 | 17.33 |
ENSRNOT00000005060
|
Mael
|
maelstrom spermatogenic transposon silencer |
| chr4_-_157408176 | 17.24 |
ENSRNOT00000021915
|
Cd4
|
Cd4 molecule |
| chr20_+_4087618 | 16.98 |
ENSRNOT00000000522
ENSRNOT00000060327 ENSRNOT00000080590 |
RT1-Db1
|
RT1 class II, locus Db1 |
| chr8_+_79054237 | 16.43 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr7_-_27552078 | 15.60 |
ENSRNOT00000059538
|
Stab2
|
stabilin 2 |
| chr2_+_138194136 | 15.40 |
ENSRNOT00000014928
|
RGD1307595
|
similar to RIKEN cDNA 1700018B24 |
| chr1_-_227175096 | 15.39 |
ENSRNOT00000054811
|
AABR07006259.1
|
|
| chr20_+_4188766 | 15.06 |
ENSRNOT00000081438
ENSRNOT00000060378 |
Tesb
|
testis specific basic protein |
| chr1_-_220848153 | 14.93 |
ENSRNOT00000037404
|
Ctsw
|
cathepsin W |
| chr15_-_42518855 | 14.75 |
ENSRNOT00000076451
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_-_111965889 | 14.72 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr1_+_141120166 | 14.46 |
ENSRNOT00000050759
|
Fanci
|
Fanconi anemia, complementation group I |
| chr10_-_57243435 | 14.44 |
ENSRNOT00000005050
|
Chrne
|
cholinergic receptor nicotinic epsilon subunit |
| chr9_+_10172832 | 14.43 |
ENSRNOT00000074555
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr7_+_40318490 | 14.34 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr7_-_117440342 | 14.23 |
ENSRNOT00000018528
|
Tssk5
|
testis-specific serine kinase 5 |
| chr15_-_44411004 | 14.12 |
ENSRNOT00000031163
|
Cdca2
|
cell division cycle associated 2 |
| chr6_-_91518996 | 14.01 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr5_-_153924896 | 13.83 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr18_+_68983545 | 13.63 |
ENSRNOT00000085317
|
Stard6
|
StAR-related lipid transfer domain containing 6 |
| chr6_+_93539271 | 13.18 |
ENSRNOT00000078791
|
Tomm20l
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr20_+_8109635 | 12.59 |
ENSRNOT00000000609
|
Armc12
|
armadillo repeat containing 12 |
| chr18_+_62174670 | 12.50 |
ENSRNOT00000025362
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr4_-_170932618 | 12.39 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr6_-_95796502 | 12.32 |
ENSRNOT00000041409
|
Six6os1
|
Six6 opposite strand transcript 1 |
| chrX_+_133227660 | 12.29 |
ENSRNOT00000004591
|
Actrt1
|
actin-related protein T1 |
| chr1_+_217039755 | 11.91 |
ENSRNOT00000091603
|
LOC102552318
|
actin-like |
| chr2_+_231884337 | 11.74 |
ENSRNOT00000014695
|
Zgrf1
|
zinc finger, GRF-type containing 1 |
| chr13_-_87847263 | 11.74 |
ENSRNOT00000003650
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr3_+_94035905 | 11.71 |
ENSRNOT00000014767
|
RGD1563222
|
similar to RIKEN cDNA A930018P22 |
| chr10_+_110346453 | 11.63 |
ENSRNOT00000054928
|
Tex19.1
|
testis expressed 19.1 |
| chr5_+_172275734 | 11.49 |
ENSRNOT00000064266
|
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr18_+_87415531 | 11.43 |
ENSRNOT00000072475
|
AABR07032888.1
|
|
| chr1_-_198486157 | 11.42 |
ENSRNOT00000022758
|
Zg16
|
zymogen granule protein 16 |
| chr4_+_157563990 | 11.40 |
ENSRNOT00000024618
|
Acrbp
|
acrosin binding protein |
| chr1_+_81260548 | 11.39 |
ENSRNOT00000026669
|
Smg9
|
SMG9 nonsense mediated mRNA decay factor |
| chr18_-_68983619 | 11.28 |
ENSRNOT00000014670
|
LOC361346
|
similar to chromosome 18 open reading frame 54 |
| chr15_+_32386816 | 11.17 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr7_+_129595192 | 10.92 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr17_-_43776460 | 10.84 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr4_-_115354795 | 10.83 |
ENSRNOT00000017691
|
Cd207
|
CD207 molecule |
| chr1_+_199019517 | 10.72 |
ENSRNOT00000025419
|
Phkg2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr5_-_59165160 | 10.55 |
ENSRNOT00000029035
|
Fam221b
|
family with sequence similarity 221, member B |
| chr4_-_70628470 | 10.36 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr14_-_17225389 | 10.31 |
ENSRNOT00000052035
|
Art3
|
ADP-ribosyltransferase 3 |
| chr10_+_74762763 | 10.25 |
ENSRNOT00000077390
|
AABR07030184.1
|
|
| chr20_+_295250 | 10.02 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chrX_-_64036217 | 10.01 |
ENSRNOT00000078311
|
AABR07038925.1
|
|
| chr12_+_25119355 | 9.99 |
ENSRNOT00000034629
|
Lat2
|
linker for activation of T cells family, member 2 |
| chr5_+_153976535 | 9.64 |
ENSRNOT00000030881
|
LOC500567
|
similar to RIKEN cDNA 1700029M20 |
| chr3_-_103745236 | 9.55 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr6_-_128690741 | 9.53 |
ENSRNOT00000035826
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr2_+_104290726 | 9.44 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr7_+_97984862 | 9.37 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr9_+_38544398 | 9.25 |
ENSRNOT00000016828
|
Prim2
|
primase (DNA) subunit 2 |
| chr12_+_12756452 | 9.20 |
ENSRNOT00000001382
ENSRNOT00000092462 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr4_+_162078905 | 9.14 |
ENSRNOT00000049620
|
LOC100910725
|
C-type lectin domain family 2 member D-related protein-like |
| chr1_-_89269930 | 9.03 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr10_-_77512032 | 9.00 |
ENSRNOT00000003295
|
Pctp
|
phosphatidylcholine transfer protein |
| chrX_-_105793306 | 8.76 |
ENSRNOT00000015806
|
Nxf2
|
nuclear RNA export factor 2 |
| chr20_+_11436267 | 8.74 |
ENSRNOT00000001631
|
Trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr20_-_13173774 | 8.64 |
ENSRNOT00000001704
|
Spatc1l
|
spermatogenesis and centriole associated 1-like |
| chr2_-_187668677 | 8.56 |
ENSRNOT00000056898
ENSRNOT00000092563 |
Tsacc
|
TSSK6 activating co-chaperone |
| chr12_+_16912249 | 8.50 |
ENSRNOT00000085936
|
Tmem184a
|
transmembrane protein 184A |
| chr9_+_81518176 | 8.43 |
ENSRNOT00000078317
ENSRNOT00000019265 ENSRNOT00000088246 ENSRNOT00000084682 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr3_-_7051953 | 8.35 |
ENSRNOT00000013473
|
RGD1306233
|
similar to hypothetical protein MGC29761 |
| chr4_+_170820594 | 8.29 |
ENSRNOT00000032568
|
LOC500354
|
similar to C030030A07Rik protein |
| chr5_+_129052043 | 8.16 |
ENSRNOT00000013459
|
LOC108348114
|
tetratricopeptide repeat protein 39A |
| chrX_+_13441558 | 7.99 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr16_+_72216326 | 7.95 |
ENSRNOT00000051363
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr12_-_51965779 | 7.89 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr6_-_127248372 | 7.86 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr3_+_19045214 | 7.79 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr3_+_122928964 | 7.77 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr20_-_29990111 | 7.71 |
ENSRNOT00000048029
|
Cdh23
|
cadherin-related 23 |
| chr20_-_12835044 | 7.69 |
ENSRNOT00000074268
|
LOC108348157
|
speriolin-like protein |
| chr2_+_211880262 | 7.56 |
ENSRNOT00000038315
|
Slc25a54
|
solute carrier family 25, member 54 |
| chr3_-_110517163 | 7.56 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr19_-_10976396 | 7.53 |
ENSRNOT00000073239
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr3_-_14571640 | 7.46 |
ENSRNOT00000060013
|
Ggta1l1
|
glycoprotein, alpha-galactosyltransferase 1-like 1 |
| chr7_+_145068286 | 7.38 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chrX_+_106230013 | 7.36 |
ENSRNOT00000067828
|
AABR07040617.1
|
|
| chr1_+_199019289 | 7.35 |
ENSRNOT00000087142
|
Phkg2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr10_+_31324512 | 7.32 |
ENSRNOT00000008559
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr19_-_37952501 | 7.29 |
ENSRNOT00000026809
|
Dpep3
|
dipeptidase 3 |
| chr1_+_221443896 | 7.25 |
ENSRNOT00000073942
|
Tmem262
|
transmembrane protein 262 |
| chr5_-_155258392 | 7.17 |
ENSRNOT00000017065
|
C1qc
|
complement C1q C chain |
| chr5_+_151343493 | 7.11 |
ENSRNOT00000077807
|
Wasf2
|
WAS protein family, member 2 |
| chr4_+_157057601 | 7.08 |
ENSRNOT00000040592
|
Zfp42l
|
Zinc finger protein 42-like |
| chr1_+_165506361 | 7.04 |
ENSRNOT00000024156
|
Ucp2
|
uncoupling protein 2 |
| chr5_+_169288871 | 6.89 |
ENSRNOT00000055466
|
Tnfrsf25
|
TNF receptor superfamily member 25 |
| chr1_-_252461461 | 6.88 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr3_+_124088157 | 6.86 |
ENSRNOT00000028876
|
Smox
|
spermine oxidase |
| chr8_-_63155825 | 6.86 |
ENSRNOT00000011820
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr14_-_21252538 | 6.83 |
ENSRNOT00000005003
|
Ambn
|
ameloblastin |
| chr1_+_146976975 | 6.82 |
ENSRNOT00000071895
|
LOC100366231
|
X-linked lymphocyte-regulated 5C-like |
| chr10_+_57064482 | 6.81 |
ENSRNOT00000046807
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr20_-_44255064 | 6.81 |
ENSRNOT00000000735
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr5_-_104920906 | 6.65 |
ENSRNOT00000071318
|
Fam154a
|
family with sequence similarity 154, member A |
| chr3_+_138974871 | 6.63 |
ENSRNOT00000012524
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr5_+_157165341 | 6.63 |
ENSRNOT00000087072
|
Pla2g2c
|
phospholipase A2, group IIC |
| chr5_+_129257429 | 6.60 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chrX_+_156227830 | 6.57 |
ENSRNOT00000083949
|
LOC690348
|
similar to ESO3 protein |
| chr15_+_27346731 | 6.56 |
ENSRNOT00000046317
|
Olr1619
|
olfactory receptor 1619 |
| chr3_+_38367556 | 6.55 |
ENSRNOT00000049144
|
RGD1559995
|
similar to developmental pluripotency associated 5 |
| chr17_-_29895386 | 6.54 |
ENSRNOT00000048757
|
Cdyl
|
chromodomain Y-like |
| chr10_-_57436368 | 6.52 |
ENSRNOT00000056608
|
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr6_-_55647665 | 6.50 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr3_+_149606940 | 6.46 |
ENSRNOT00000018512
|
Bpifa3
|
BPI fold containing family A, member 3 |
| chr5_-_127273656 | 6.45 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr20_+_4967194 | 6.41 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_-_21761487 | 6.37 |
ENSRNOT00000082910
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr16_-_31301880 | 6.30 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr13_-_79899479 | 6.27 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chrX_-_156540733 | 6.26 |
ENSRNOT00000091029
|
LOC108348261
|
transketolase-like protein 1 |
| chr19_+_55381565 | 6.23 |
ENSRNOT00000018923
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr16_+_6434468 | 6.21 |
ENSRNOT00000012906
|
LOC108348453
|
calsequestrin-1-like |
| chr6_-_135112775 | 6.16 |
ENSRNOT00000086310
|
LOC103692716
|
heat shock protein HSP 90-alpha |
| chr1_-_12952906 | 6.15 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr20_-_4921348 | 6.14 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr7_+_70612103 | 6.12 |
ENSRNOT00000057833
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr4_-_161757447 | 6.05 |
ENSRNOT00000008737
|
Fkbp4
|
FK506 binding protein 4 |
| chr5_-_62187930 | 6.03 |
ENSRNOT00000011787
|
Coro2a
|
coronin 2A |
| chr3_+_19690016 | 5.87 |
ENSRNOT00000085460
|
AABR07051707.1
|
|
| chr10_-_7029136 | 5.84 |
ENSRNOT00000091942
|
AC129395.1
|
|
| chrX_+_105937118 | 5.82 |
ENSRNOT00000045591
|
Pramel
|
preferentially expressed antigen in melanoma-like |
| chr5_+_160095427 | 5.81 |
ENSRNOT00000077609
|
AABR07050298.1
|
|
| chrX_+_106002416 | 5.71 |
ENSRNOT00000052196
|
LOC100910698
|
leucine-rich repeat-containing protein PRAME-like |
| chr5_+_5866897 | 5.67 |
ENSRNOT00000011940
|
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr4_-_162230859 | 5.66 |
ENSRNOT00000042872
|
LOC689757
|
similar to osteoclast inhibitory lectin |
| chr10_-_56488723 | 5.63 |
ENSRNOT00000065524
|
Tmem95
|
transmembrane protein 95 |
| chr8_+_22423467 | 5.59 |
ENSRNOT00000085320
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr1_-_99985422 | 5.49 |
ENSRNOT00000025701
|
Klk1c2
|
kallikrein 1-related peptidase C2 |
| chr17_+_15762030 | 5.45 |
ENSRNOT00000089310
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr20_+_3148665 | 5.39 |
ENSRNOT00000086026
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr1_+_81643816 | 5.32 |
ENSRNOT00000027214
|
LOC103689942
|
carcinoembryonic antigen-related cell adhesion molecule 1-like |
| chr1_-_211196868 | 5.32 |
ENSRNOT00000022714
|
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr1_+_89162639 | 5.31 |
ENSRNOT00000028508
|
Atp4a
|
ATPase H+/K+ transporting alpha subunit |
| chr1_-_190914610 | 5.22 |
ENSRNOT00000023189
|
Cdr2
|
cerebellar degeneration-related protein 2 |
| chr8_-_12355091 | 5.21 |
ENSRNOT00000009318
|
Cep57
|
centrosomal protein 57 |
| chr8_+_2604962 | 5.21 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr20_+_3149114 | 5.21 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr3_+_2877293 | 5.16 |
ENSRNOT00000061855
|
Lcn5
|
lipocalin 5 |
| chr4_+_102403451 | 5.16 |
ENSRNOT00000071605
|
AABR07060995.1
|
|
| chr9_-_15410943 | 5.15 |
ENSRNOT00000074217
|
Ccnd3
|
cyclin D3 |
| chr4_-_183426439 | 5.10 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chr5_-_48504511 | 5.08 |
ENSRNOT00000010271
|
Pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_-_140334912 | 5.06 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr8_+_118744229 | 4.86 |
ENSRNOT00000028364
|
Kif9
|
kinesin family member 9 |
| chr1_+_279798187 | 4.83 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr12_-_31323810 | 4.78 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr5_+_135574172 | 4.75 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr16_-_20807070 | 4.74 |
ENSRNOT00000072536
|
Comp
|
cartilage oligomeric matrix protein |
| chr20_-_32133431 | 4.74 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr2_-_84678790 | 4.74 |
ENSRNOT00000015886
|
Cct5
|
chaperonin containing TCP1 subunit 5 |
| chr9_-_65693822 | 4.73 |
ENSRNOT00000038431
|
Als2cr12
|
amyotrophic lateral sclerosis 2 chromosome region, candidate 12 |
| chr17_+_5311274 | 4.70 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr3_+_155119042 | 4.69 |
ENSRNOT00000032943
|
Actr5
|
ARP5 actin-related protein 5 homolog |
| chr14_-_81339526 | 4.67 |
ENSRNOT00000015894
|
Grk4
|
G protein-coupled receptor kinase 4 |
| chr8_+_69121682 | 4.57 |
ENSRNOT00000013461
|
Rpl4
|
ribosomal protein L4 |
| chr1_+_82452469 | 4.57 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr14_+_107767392 | 4.53 |
ENSRNOT00000012847
|
Cct4
|
chaperonin containing TCP1 subunit 4 |
| chr11_-_87858107 | 4.52 |
ENSRNOT00000002550
|
Snap29
|
synaptosomal-associated protein 29 |
| chr5_+_162385173 | 4.40 |
ENSRNOT00000048232
|
Pramef12
|
PRAME family member 12 |
| chrX_-_112159458 | 4.39 |
ENSRNOT00000087403
|
Tex13b
|
testis expressed 13B |
| chr5_-_137281277 | 4.37 |
ENSRNOT00000093752
ENSRNOT00000093708 ENSRNOT00000093301 ENSRNOT00000093502 ENSRNOT00000034762 |
Mpl
|
MPL proto-oncogene, thrombopoietin receptor |
| chr7_+_72772440 | 4.29 |
ENSRNOT00000009989
|
Mtdh
|
metadherin |
| chr11_+_70034139 | 4.27 |
ENSRNOT00000002450
|
Umps
|
uridine monophosphate synthetase |
| chr11_-_61530567 | 4.24 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr20_-_5037022 | 4.23 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr15_+_52234563 | 4.21 |
ENSRNOT00000015169
|
Reep4
|
receptor accessory protein 4 |
| chr1_-_174119815 | 4.19 |
ENSRNOT00000019368
|
Trim66
|
tripartite motif-containing 66 |
| chr3_+_114102875 | 4.18 |
ENSRNOT00000023209
|
Trim69
|
tripartite motif-containing 69 |
| chr20_+_21316826 | 4.17 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr1_-_145931583 | 4.15 |
ENSRNOT00000016433
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr7_+_40217991 | 4.11 |
ENSRNOT00000085684
|
Cep290
|
centrosomal protein 290 |
| chr8_+_22423890 | 4.09 |
ENSRNOT00000009354
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr2_+_187708234 | 4.07 |
ENSRNOT00000026514
|
Smg5
|
SMG5 nonsense mediated mRNA decay factor |
| chr15_+_87886783 | 4.05 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr7_-_143392777 | 4.05 |
ENSRNOT00000086504
ENSRNOT00000038105 |
Krt72
|
keratin 72 |
| chr1_+_147080783 | 4.03 |
ENSRNOT00000065898
|
LOC100911148
|
PWWP domain-containing protein MUM1-like |
| chr16_-_75004617 | 3.99 |
ENSRNOT00000080407
|
Fam90a1a
|
family with sequence similarity 90, member A1A |
| chr7_-_107203897 | 3.97 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr7_-_59763219 | 3.94 |
ENSRNOT00000041210
|
RGD1564883
|
similar to 60S ribosomal protein L12 |
| chr11_-_27141881 | 3.93 |
ENSRNOT00000002169
|
Cct8
|
chaperonin containing TCP1 subunit 8 |
| chr11_-_36533073 | 3.91 |
ENSRNOT00000033486
|
Lca5l
|
LCA5L, lebercilin like |
| chr15_+_57709043 | 3.88 |
ENSRNOT00000030579
|
Erich6b
|
glutamate-rich 6B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 24.1 | GO:0070103 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 6.4 | 32.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 5.7 | 17.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 4.9 | 29.2 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 4.2 | 12.5 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 4.1 | 12.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 4.0 | 7.9 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) |
| 3.7 | 14.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 2.8 | 17.0 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 2.8 | 14.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 2.5 | 7.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 2.3 | 9.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 2.1 | 8.5 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 2.1 | 8.4 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 2.1 | 6.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) regulation of DNA replication origin binding(GO:1902595) |
| 1.9 | 7.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 1.7 | 8.7 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 1.7 | 13.5 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 1.7 | 30.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 1.7 | 11.6 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 1.6 | 4.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.5 | 9.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.5 | 14.6 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 1.5 | 21.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 1.4 | 10.0 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 1.3 | 5.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.3 | 8.9 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 1.3 | 10.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 1.2 | 3.7 | GO:0043324 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 1.2 | 17.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.2 | 6.2 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 1.2 | 20.9 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 1.2 | 9.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.1 | 6.9 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 1.1 | 4.6 | GO:0045006 | DNA deamination(GO:0045006) |
| 1.1 | 3.4 | GO:0036115 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 1.1 | 12.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.1 | 4.5 | GO:0016240 | autophagosome docking(GO:0016240) |
| 1.1 | 3.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 1.1 | 4.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.0 | 14.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.9 | 13.8 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.9 | 9.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.9 | 16.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.9 | 9.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.9 | 7.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.8 | 7.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.8 | 23.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.8 | 2.5 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.8 | 5.4 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.8 | 2.3 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.7 | 5.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.7 | 2.2 | GO:0021508 | floor plate formation(GO:0021508) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.7 | 4.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.7 | 19.0 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.6 | 20.5 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.6 | 8.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.6 | 1.9 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.6 | 8.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.6 | 2.9 | GO:0036233 | glycine import(GO:0036233) |
| 0.6 | 7.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.6 | 1.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.5 | 14.5 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.5 | 4.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.5 | 4.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.5 | 2.6 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.5 | 2.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 4.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.5 | 2.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.5 | 16.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.5 | 11.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.5 | 4.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.5 | 1.9 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.5 | 2.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.5 | 1.8 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.5 | 19.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.5 | 3.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.4 | 4.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 5.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.4 | 1.7 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.4 | 10.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.9 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.4 | 10.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.4 | 14.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.4 | 7.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.4 | 2.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.3 | 3.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 3.0 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.3 | 5.3 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.3 | 1.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.0 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.3 | 5.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 4.8 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.3 | 1.3 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) cellular response to interleukin-18(GO:0071351) |
| 0.3 | 1.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 3.9 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.3 | 1.5 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 7.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.3 | 3.7 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.3 | 3.3 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.3 | 7.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 14.1 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.3 | 13.7 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.2 | 0.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.2 | 4.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 5.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 0.5 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.2 | 1.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 3.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 12.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 0.6 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 7.3 | GO:0007140 | male meiosis(GO:0007140) |
| 0.2 | 4.9 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.2 | 0.9 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.2 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 4.0 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 1.7 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 3.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.7 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.1 | 2.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 14.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 3.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 9.3 | GO:0007127 | meiosis I(GO:0007127) |
| 0.1 | 0.5 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 | 4.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 8.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 2.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 5.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.6 | GO:0090367 | regulation of mRNA modification(GO:0090365) negative regulation of mRNA modification(GO:0090367) |
| 0.1 | 12.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 29.1 | GO:0070489 | T cell activation(GO:0042110) T cell aggregation(GO:0070489) |
| 0.1 | 3.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 7.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 3.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 4.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 3.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 5.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 3.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 2.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 2.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 10.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 1.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.5 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 2.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 2.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.3 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 1.8 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 2.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 29.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 2.9 | 17.3 | GO:0071547 | piP-body(GO:0071547) |
| 2.8 | 14.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 2.5 | 7.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 2.3 | 11.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 2.3 | 18.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 2.0 | 9.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.6 | 11.4 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.6 | 14.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.6 | 7.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.4 | 10.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.4 | 8.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.2 | 4.8 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 1.2 | 9.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.1 | 4.5 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.1 | 17.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.0 | 5.2 | GO:0072558 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 1.0 | 13.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.0 | 7.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.9 | 3.8 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.9 | 12.3 | GO:0000801 | central element(GO:0000801) |
| 0.9 | 18.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.8 | 6.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.8 | 3.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.7 | 14.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.7 | 2.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.7 | 2.6 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.6 | 4.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.6 | 1.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.6 | 7.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 14.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 8.9 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.5 | 8.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.5 | 3.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 11.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 7.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 40.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 4.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.4 | 2.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.3 | 2.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 5.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 4.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 2.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 0.9 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.3 | 4.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 1.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.3 | 41.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.3 | 2.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) chromaffin granule(GO:0042583) neurofibrillary tangle(GO:0097418) |
| 0.2 | 21.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 2.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 4.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 2.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 0.6 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.2 | 6.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 5.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 10.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 1.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 4.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 1.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 19.8 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 8.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 4.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 2.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 4.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 10.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 5.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.9 | GO:0071004 | U1 snRNP(GO:0005685) U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 6.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 33.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 7.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 6.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 6.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 5.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 3.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 18.4 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 3.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 13.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 4.3 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 19.8 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.9 | 17.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.6 | 7.9 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.3 | 6.9 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 2.3 | 18.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 2.1 | 14.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.9 | 7.5 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 1.8 | 7.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.7 | 6.8 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 1.6 | 19.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.4 | 8.4 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 1.4 | 20.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 1.3 | 5.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 1.3 | 9.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.3 | 10.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 1.2 | 6.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.2 | 15.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 1.2 | 4.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.0 | 8.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 1.0 | 9.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.9 | 17.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.9 | 18.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.9 | 12.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.9 | 6.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.9 | 8.7 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.9 | 3.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.8 | 14.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.8 | 7.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.8 | 9.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.8 | 12.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.8 | 4.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.7 | 6.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.7 | 7.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 8.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.7 | 2.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.7 | 16.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.6 | 1.9 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.6 | 2.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.6 | 9.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 10.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 1.7 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.4 | 20.3 | GO:0032934 | sterol binding(GO:0032934) |
| 0.4 | 2.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 5.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 8.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 4.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 4.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 7.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 1.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.4 | 4.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 12.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 1.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.3 | 10.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 6.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 6.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 3.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 2.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 3.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.3 | 2.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 3.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 4.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 2.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 18.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 2.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 1.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 5.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 5.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 25.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 6.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 2.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 6.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 2.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 3.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 13.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 4.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 15.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 6.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 20.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 4.6 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 2.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 5.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.5 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 7.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 7.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 3.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 3.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.2 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 1.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 5.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 7.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 5.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 10.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 14.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 16.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 6.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 45.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.6 | 26.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.6 | 15.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 7.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 19.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.4 | 30.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 18.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 15.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 5.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 18.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 13.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.3 | 3.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 12.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 10.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 5.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 3.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 7.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 46.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 2.1 | 20.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.2 | 14.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 1.2 | 30.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 1.0 | 24.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.9 | 7.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.8 | 29.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.8 | 7.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.7 | 13.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.7 | 14.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.7 | 9.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.7 | 7.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 19.8 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.6 | 7.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 10.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 9.6 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.4 | 5.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 20.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 6.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 4.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 3.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.3 | 11.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 7.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 10.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 5.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 23.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 10.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 4.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 2.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 9.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 5.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 4.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 4.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |