Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
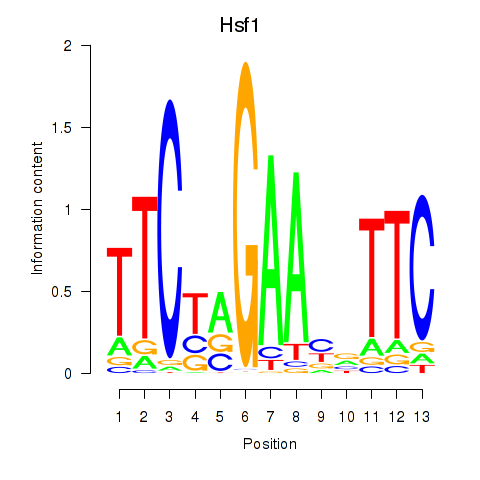
Results for Hsf1
Z-value: 0.79
Transcription factors associated with Hsf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf1
|
ENSRNOG00000021732 | heat shock transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf1 | rn6_v1_chr7_+_117538523_117538523 | 0.12 | 2.7e-02 | Click! |
Activity profile of Hsf1 motif
Sorted Z-values of Hsf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_17225389 | 33.85 |
ENSRNOT00000052035
|
Art3
|
ADP-ribosyltransferase 3 |
| chr10_+_70262361 | 29.86 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr6_-_94980004 | 19.56 |
ENSRNOT00000006373
|
Rtn1
|
reticulon 1 |
| chr6_-_127248372 | 17.78 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr8_+_70603249 | 15.35 |
ENSRNOT00000067016
ENSRNOT00000072486 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr5_+_58393233 | 13.46 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr4_-_161757447 | 12.19 |
ENSRNOT00000008737
|
Fkbp4
|
FK506 binding protein 4 |
| chr7_+_11737293 | 11.51 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr9_+_10172832 | 10.53 |
ENSRNOT00000074555
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr8_+_55178289 | 10.35 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr14_-_43143973 | 10.29 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr20_+_8109635 | 9.92 |
ENSRNOT00000000609
|
Armc12
|
armadillo repeat containing 12 |
| chr1_-_222293148 | 9.66 |
ENSRNOT00000028743
|
Stip1
|
stress-induced phosphoprotein 1 |
| chr1_-_12952906 | 9.47 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr8_-_111965889 | 9.05 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr7_+_129595192 | 8.98 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr3_+_112228720 | 8.81 |
ENSRNOT00000079079
|
Capn3
|
calpain 3 |
| chr11_-_61530567 | 7.75 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr10_+_5930298 | 7.65 |
ENSRNOT00000044626
|
Grin2a
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr9_+_73334618 | 7.55 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chrX_+_45213228 | 7.54 |
ENSRNOT00000088538
|
LOC103690319
|
odorant-binding protein |
| chr4_+_89079014 | 6.96 |
ENSRNOT00000087451
|
Herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr7_-_116106368 | 6.63 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr20_+_4188766 | 6.61 |
ENSRNOT00000081438
ENSRNOT00000060378 |
Tesb
|
testis specific basic protein |
| chr7_-_117440342 | 6.45 |
ENSRNOT00000018528
|
Tssk5
|
testis-specific serine kinase 5 |
| chr2_-_173087648 | 6.43 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr18_-_1946840 | 6.34 |
ENSRNOT00000041878
|
Abhd3
|
abhydrolase domain containing 3 |
| chr2_+_84678948 | 6.20 |
ENSRNOT00000046325
|
Fam173b
|
family with sequence similarity 173, member B |
| chr11_-_81379640 | 5.85 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr8_+_17421557 | 5.69 |
ENSRNOT00000030569
|
Chordc1
|
cysteine and histidine rich domain containing 1 |
| chr1_-_216971183 | 5.60 |
ENSRNOT00000077911
|
Mrgpre
|
MAS related GPR family member E |
| chr3_+_38367556 | 5.33 |
ENSRNOT00000049144
|
RGD1559995
|
similar to developmental pluripotency associated 5 |
| chrX_-_83864150 | 5.31 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr11_-_81379871 | 5.11 |
ENSRNOT00000089294
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr11_+_84396033 | 4.95 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chrX_-_45284341 | 4.94 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr16_+_20336638 | 4.85 |
ENSRNOT00000080467
|
Kcnn1
|
potassium calcium-activated channel subfamily N member 1 |
| chr2_-_187668677 | 4.75 |
ENSRNOT00000056898
ENSRNOT00000092563 |
Tsacc
|
TSSK6 activating co-chaperone |
| chr8_+_98745310 | 4.64 |
ENSRNOT00000019938
|
Zic4
|
Zic family member 4 |
| chr1_-_48033343 | 4.63 |
ENSRNOT00000019531
ENSRNOT00000076422 |
Tcp1
|
t-complex 1 |
| chr10_-_105787803 | 4.60 |
ENSRNOT00000064671
|
Jmjd6
|
jumonji domain containing 6 |
| chr10_+_105787935 | 4.54 |
ENSRNOT00000000266
|
Mettl23
|
methyltransferase like 23 |
| chr11_-_28672956 | 4.54 |
ENSRNOT00000002137
|
LOC100363136
|
hypothetical protein LOC100363136 |
| chr15_+_29208606 | 4.46 |
ENSRNOT00000073722
ENSRNOT00000086054 |
LOC100911980
|
uncharacterized LOC100911980 |
| chrX_-_112328642 | 4.35 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr5_+_169357517 | 4.35 |
ENSRNOT00000000079
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr6_+_93539271 | 4.34 |
ENSRNOT00000078791
|
Tomm20l
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr15_+_87886783 | 4.33 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr2_+_80948658 | 4.05 |
ENSRNOT00000074689
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr7_+_40318490 | 3.74 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr14_+_7169519 | 3.44 |
ENSRNOT00000003026
|
Klhl8
|
kelch-like family member 8 |
| chr3_+_122928964 | 3.44 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr11_-_27141881 | 3.21 |
ENSRNOT00000002169
|
Cct8
|
chaperonin containing TCP1 subunit 8 |
| chr7_-_69982592 | 3.15 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr11_+_28692708 | 3.12 |
ENSRNOT00000002135
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr1_+_221443896 | 3.10 |
ENSRNOT00000073942
|
Tmem262
|
transmembrane protein 262 |
| chr5_+_160095427 | 2.98 |
ENSRNOT00000077609
|
AABR07050298.1
|
|
| chr13_-_79899479 | 2.96 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chr4_-_154044493 | 2.90 |
ENSRNOT00000076318
|
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr1_-_89045586 | 2.83 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr3_-_14571640 | 2.76 |
ENSRNOT00000060013
|
Ggta1l1
|
glycoprotein, alpha-galactosyltransferase 1-like 1 |
| chr15_+_32386816 | 2.45 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr3_+_94035905 | 2.44 |
ENSRNOT00000014767
|
RGD1563222
|
similar to RIKEN cDNA A930018P22 |
| chr12_-_6956914 | 2.42 |
ENSRNOT00000072129
ENSRNOT00000001210 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr10_-_56488723 | 2.41 |
ENSRNOT00000065524
|
Tmem95
|
transmembrane protein 95 |
| chr19_+_10167797 | 2.25 |
ENSRNOT00000085440
|
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chrX_+_15321713 | 2.25 |
ENSRNOT00000071835
|
LOC108348138
|
GTPase ERas |
| chr7_-_125497691 | 2.24 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chr10_+_86564928 | 2.16 |
ENSRNOT00000039892
|
Lrrc3c
|
leucine rich repeat containing 3C |
| chr11_+_28739219 | 1.97 |
ENSRNOT00000061620
|
Krtap13-2
|
keratin associated protein 13-2 |
| chr1_+_192025357 | 1.90 |
ENSRNOT00000025072
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr8_-_58293102 | 1.79 |
ENSRNOT00000011913
|
Rab39a
|
RAB39A, member RAS oncogene family |
| chr2_-_84678790 | 1.74 |
ENSRNOT00000015886
|
Cct5
|
chaperonin containing TCP1 subunit 5 |
| chr12_-_23841049 | 1.43 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr20_+_4118461 | 1.35 |
ENSRNOT00000083085
|
RT1-Db2
|
RT1 class II, locus Db2 |
| chr3_+_154043873 | 1.33 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr20_+_3149114 | 1.27 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr12_+_38274297 | 1.08 |
ENSRNOT00000087905
ENSRNOT00000057788 |
Rsrc2
|
arginine and serine rich coiled-coil 2 |
| chr12_+_12756452 | 1.06 |
ENSRNOT00000001382
ENSRNOT00000092462 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr12_-_31323810 | 0.94 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr14_-_108273938 | 0.88 |
ENSRNOT00000084642
|
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr2_+_178354890 | 0.87 |
ENSRNOT00000037890
|
Ppid
|
peptidylprolyl isomerase D |
| chr12_-_30501860 | 0.50 |
ENSRNOT00000001227
|
Cct6a
|
chaperonin containing TCP1 subunit 6A |
| chr17_+_36771639 | 0.46 |
ENSRNOT00000015147
|
Gpr50
|
G protein-coupled receptor 50 |
| chr14_+_88543630 | 0.46 |
ENSRNOT00000060515
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr8_+_132828091 | 0.46 |
ENSRNOT00000008269
|
Ccr9
|
C-C motif chemokine receptor 9 |
| chr1_-_192025350 | 0.40 |
ENSRNOT00000071946
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr20_-_44255064 | 0.29 |
ENSRNOT00000000735
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr5_-_151768123 | 0.23 |
ENSRNOT00000079380
|
Nudc
|
nuclear distribution C, dynein complex regulator |
| chr1_+_105094411 | 0.20 |
ENSRNOT00000036258
|
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr18_-_29611212 | 0.17 |
ENSRNOT00000022685
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chrX_-_45187149 | 0.16 |
ENSRNOT00000080431
|
Pbsn
|
probasin |
| chr9_-_61690956 | 0.16 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr1_+_80135391 | 0.13 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr2_+_187668796 | 0.13 |
ENSRNOT00000025824
|
Cct3
|
chaperonin containing TCP1 subunit 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 2.5 | 7.6 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.9 | 5.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.5 | 4.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.4 | 4.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 1.3 | 10.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.3 | 10.1 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 1.2 | 33.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 1.2 | 4.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 1.1 | 10.3 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.9 | 7.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.7 | 12.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.6 | 9.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.6 | 2.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.5 | 28.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.3 | 1.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.3 | 4.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.3 | 2.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 11.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 6.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 17.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 2.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 7.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 13.5 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 10.5 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 3.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.1 | 5.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 1.4 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 2.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.5 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 11.5 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 4.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 4.9 | GO:0032868 | response to insulin(GO:0032868) |
| 0.0 | 1.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 2.1 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 14.8 | GO:0030182 | neuron differentiation(GO:0030182) |
| 0.0 | 4.5 | GO:0032259 | methylation(GO:0032259) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.3 | 11.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.1 | 7.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.8 | 7.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.8 | 10.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.7 | 7.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.7 | 4.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.5 | 19.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 4.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.3 | 12.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 0.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 2.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 10.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 0.5 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.1 | 8.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 9.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 5.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 68.2 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 33.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 2.6 | 10.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.4 | 12.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.2 | 4.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 1.1 | 4.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.1 | 6.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.9 | 36.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.9 | 19.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.9 | 10.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.8 | 7.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.7 | 4.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.7 | 2.8 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.6 | 8.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.6 | 7.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 11.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 7.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 9.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 2.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 0.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 2.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 11.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 13.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 15.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 4.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 17.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 9.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 4.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 4.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 4.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 6.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 7.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 12.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 7.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.5 | 11.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 7.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 2.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 4.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 5.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 9.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 15.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 4.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |