Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxd9
Z-value: 0.55
Transcription factors associated with Hoxd9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd9
|
ENSRNOG00000001580 | homeo box D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd9 | rn6_v1_chr3_+_61620192_61620192 | 0.35 | 1.8e-10 | Click! |
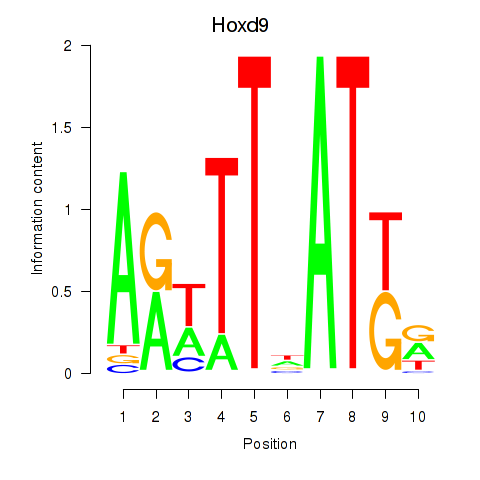
Activity profile of Hoxd9 motif
Sorted Z-values of Hoxd9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22517774 | 31.61 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr11_-_81444375 | 22.55 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr14_+_22375955 | 19.72 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr14_-_19132208 | 17.81 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr5_-_147784311 | 17.79 |
ENSRNOT00000074172
|
Fam167b
|
family with sequence similarity 167, member B |
| chr7_-_73270308 | 16.92 |
ENSRNOT00000007430
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr2_+_252452269 | 16.58 |
ENSRNOT00000021970
|
Uox
|
urate oxidase |
| chr13_+_56598957 | 15.43 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr9_+_95295701 | 13.25 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr4_-_51199570 | 12.33 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr8_+_59507990 | 12.22 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chr2_+_54466280 | 11.33 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr10_-_47997796 | 9.28 |
ENSRNOT00000078422
|
Slc5a10
|
solute carrier family 5 member 10 |
| chr16_+_50152008 | 7.71 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr4_-_176679815 | 7.23 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr2_-_158156444 | 7.06 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr9_-_79545024 | 6.96 |
ENSRNOT00000021152
|
Mreg
|
melanoregulin |
| chr1_+_21613148 | 6.64 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_+_141218095 | 6.40 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr13_-_56462834 | 6.25 |
ENSRNOT00000032122
|
Crb1
|
crumbs 1, cell polarity complex component |
| chr3_-_77483322 | 6.15 |
ENSRNOT00000008077
|
Olr661
|
olfactory receptor 661 |
| chr1_+_128606770 | 5.93 |
ENSRNOT00000073355
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_+_55812747 | 5.85 |
ENSRNOT00000008106
|
Sostdc1
|
sclerostin domain containing 1 |
| chr3_-_77227036 | 4.90 |
ENSRNOT00000072638
|
LOC103691843
|
olfactory receptor 4P4-like |
| chr2_-_178612470 | 4.84 |
ENSRNOT00000013499
|
Tmem144
|
transmembrane protein 144 |
| chr10_-_67478848 | 4.81 |
ENSRNOT00000005325
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr4_+_33890349 | 4.68 |
ENSRNOT00000078680
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr12_-_6078411 | 4.55 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr13_+_85818427 | 4.47 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr3_+_121490812 | 4.41 |
ENSRNOT00000023913
|
AABR07053707.1
|
|
| chrX_-_115073890 | 3.91 |
ENSRNOT00000006638
|
Capn6
|
calpain 6 |
| chr15_+_12363377 | 3.81 |
ENSRNOT00000008805
|
Olr1607
|
olfactory receptor 1607 |
| chr17_+_45801528 | 3.70 |
ENSRNOT00000089221
|
Olr1664
|
olfactory receptor 1664 |
| chr15_-_9086282 | 3.59 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr4_-_183544850 | 2.96 |
ENSRNOT00000071407
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr14_+_12218553 | 2.90 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr17_-_51912496 | 2.67 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr1_-_141471010 | 2.67 |
ENSRNOT00000082164
|
Plin1
|
perilipin 1 |
| chr3_+_138508899 | 2.50 |
ENSRNOT00000009437
|
Kat14
|
lysine acetyltransferase 14 |
| chr15_+_28894627 | 2.45 |
ENSRNOT00000017795
|
Olr1643
|
olfactory receptor 1643 |
| chr2_-_252451999 | 2.39 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr3_-_73523043 | 2.38 |
ENSRNOT00000012959
|
Olr484
|
olfactory receptor 484 |
| chr19_+_683484 | 2.28 |
ENSRNOT00000090332
|
Terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr3_-_77826506 | 2.28 |
ENSRNOT00000071385
|
Olr675
|
olfactory receptor 675 |
| chr4_-_13878126 | 2.27 |
ENSRNOT00000007032
|
Gnat3
|
G protein subunit alpha transducin 3 |
| chr17_+_81280898 | 2.26 |
ENSRNOT00000057785
|
Tmem236
|
transmembrane protein 236 |
| chr2_+_35310188 | 2.24 |
ENSRNOT00000073523
|
LOC100910145
|
olfactory receptor 147-like |
| chr2_+_22910236 | 2.19 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_172813394 | 2.17 |
ENSRNOT00000090059
|
Olr272
|
olfactory receptor 272 |
| chr3_-_75576520 | 2.13 |
ENSRNOT00000083330
|
Olr567
|
olfactory receptor 567 |
| chr20_-_35342 | 2.11 |
ENSRNOT00000048277
|
LOC100910263
|
olfactory receptor 2B11-like |
| chr3_-_102996314 | 2.06 |
ENSRNOT00000046265
|
Olr774
|
olfactory receptor 774 |
| chr9_-_18371856 | 2.04 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr8_+_41252456 | 2.01 |
ENSRNOT00000071683
|
Olr1220
|
olfactory receptor 1220 |
| chr13_+_26463404 | 2.00 |
ENSRNOT00000061779
|
LOC103690041
|
serpin B12-like |
| chr3_+_75970593 | 1.99 |
ENSRNOT00000075834
|
Olr592
|
olfactory receptor 592 |
| chr14_+_44413636 | 1.98 |
ENSRNOT00000003552
|
Smim14
|
small integral membrane protein 14 |
| chr3_-_78235760 | 1.96 |
ENSRNOT00000008435
|
Olr698
|
olfactory receptor 698 |
| chr1_+_51619875 | 1.85 |
ENSRNOT00000023319
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6 |
| chr15_-_36321572 | 1.80 |
ENSRNOT00000073934
|
AABR07018016.1
|
|
| chr2_+_197715761 | 1.74 |
ENSRNOT00000083069
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr2_-_29248174 | 1.73 |
ENSRNOT00000071167
|
LOC684444
|
similar to HIStone family member (his-41) |
| chrM_+_3904 | 1.72 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_+_277355619 | 1.71 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr3_-_78016906 | 1.66 |
ENSRNOT00000089048
|
Olr687
|
olfactory receptor 687 |
| chr9_+_111233147 | 1.63 |
ENSRNOT00000056439
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr14_-_16903242 | 1.62 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chrX_+_29520263 | 1.60 |
ENSRNOT00000037047
|
Saxo1
|
stabilizer of axonemal microtubules 1 |
| chr15_+_35889080 | 1.60 |
ENSRNOT00000075355
|
LOC100910640
|
olfactory receptor 144-like |
| chr14_+_23330933 | 1.53 |
ENSRNOT00000088552
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr1_+_229755649 | 1.48 |
ENSRNOT00000046384
|
Olr340
|
olfactory receptor 340 |
| chr8_+_20230082 | 1.48 |
ENSRNOT00000044463
|
Olr1165
|
olfactory receptor 1165 |
| chr16_-_24951612 | 1.45 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr5_-_59279210 | 1.44 |
ENSRNOT00000041134
|
Olr837
|
olfactory receptor 837 |
| chr4_-_165774126 | 1.43 |
ENSRNOT00000007453
|
Tas2r107
|
taste receptor, type 2, member 107 |
| chr2_+_46203776 | 1.42 |
ENSRNOT00000074056
|
LOC108350228
|
olfactory receptor 147-like |
| chr1_-_171045419 | 1.41 |
ENSRNOT00000047239
|
Olr224
|
olfactory receptor 224 |
| chr3_-_73636542 | 1.39 |
ENSRNOT00000074870
|
LOC100909831
|
olfactory receptor 8K3-like |
| chr2_-_191294374 | 1.39 |
ENSRNOT00000067469
|
RGD1562234
|
similar to S100 calcium-binding protein, ventral prostate |
| chr1_-_60634546 | 1.38 |
ENSRNOT00000051451
|
Vom1r15
|
vomeronasal 1 receptor 15 |
| chr13_+_26903052 | 1.37 |
ENSRNOT00000003625
|
Serpinb5
|
serpin family B member 5 |
| chr1_+_168341595 | 1.37 |
ENSRNOT00000048411
|
Olr84
|
olfactory receptor 84 |
| chr1_+_169636914 | 1.37 |
ENSRNOT00000074466
|
Olr158
|
olfactory receptor 158 |
| chr8_+_41365984 | 1.35 |
ENSRNOT00000074250
|
Olr1228
|
olfactory receptor 1228 |
| chr8_-_14880644 | 1.29 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr3_-_78271875 | 1.28 |
ENSRNOT00000008475
|
Olr701
|
olfactory receptor 701 |
| chr7_+_13673934 | 1.26 |
ENSRNOT00000048564
|
Olr1085
|
olfactory receptor 1085 |
| chr18_+_29191731 | 1.26 |
ENSRNOT00000068085
ENSRNOT00000025495 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr2_-_192812008 | 1.26 |
ENSRNOT00000080554
|
Lce6a
|
late cornified envelope 6A |
| chr17_+_89452814 | 1.22 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr18_+_81821127 | 1.21 |
ENSRNOT00000058199
|
Fbxo15
|
F-box protein 15 |
| chr3_-_73124644 | 1.14 |
ENSRNOT00000012711
|
Olr455
|
olfactory receptor 455 |
| chr9_+_12114977 | 1.14 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr8_+_19548488 | 1.13 |
ENSRNOT00000008605
|
Olr1148
|
olfactory receptor 1148 |
| chr11_-_61698595 | 1.12 |
ENSRNOT00000093445
|
Ccdc191
|
coiled-coil domain containing 191 |
| chr3_-_78362120 | 1.07 |
ENSRNOT00000051799
|
Olr714
|
olfactory receptor 714 |
| chr1_-_168644038 | 1.07 |
ENSRNOT00000021304
|
Olr109
|
olfactory receptor 109 |
| chr18_+_31574821 | 1.06 |
ENSRNOT00000018499
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr1_+_172348583 | 0.99 |
ENSRNOT00000041144
|
Olr250
|
olfactory receptor 250 |
| chr1_-_22404002 | 0.99 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr4_-_101393329 | 0.98 |
ENSRNOT00000007636
|
RGD1562515
|
similar to RIKEN cDNA 4931417E11 |
| chr19_-_56983807 | 0.97 |
ENSRNOT00000043529
|
Cox6c-ps1
|
cytochrome c oxidase subunit VIc, pseudogene |
| chrX_+_32495809 | 0.95 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr18_+_40596166 | 0.95 |
ENSRNOT00000087984
|
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr1_-_200548317 | 0.94 |
ENSRNOT00000088610
|
AABR07005806.1
|
|
| chr2_-_35416967 | 0.92 |
ENSRNOT00000083497
|
Olr1274
|
olfactory receptor 1274 |
| chr4_+_72670543 | 0.92 |
ENSRNOT00000044838
|
Olr821
|
olfactory receptor 821 |
| chr2_+_237755673 | 0.87 |
ENSRNOT00000083937
|
Tbck
|
TBC1 domain containing kinase |
| chr5_-_69179068 | 0.86 |
ENSRNOT00000075727
|
LOC103690011
|
olfactory receptor 13C3-like |
| chr3_-_76696107 | 0.84 |
ENSRNOT00000044692
|
Olr629
|
olfactory receptor 629 |
| chr3_+_76842898 | 0.82 |
ENSRNOT00000085537
|
Olr636
|
olfactory receptor 636 |
| chr8_+_41302631 | 0.82 |
ENSRNOT00000076963
|
LOC100910822
|
olfactory receptor 143-like |
| chr3_-_78717401 | 0.81 |
ENSRNOT00000008676
|
Olr720
|
olfactory receptor 720 |
| chr3_+_75582317 | 0.80 |
ENSRNOT00000079367
|
Olr569
|
olfactory receptor 569 |
| chr7_+_122160171 | 0.76 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr5_-_110706656 | 0.75 |
ENSRNOT00000033781
|
Izumo3
|
IZUMO family member 3 |
| chr3_-_74503106 | 0.75 |
ENSRNOT00000057508
|
Olr529
|
olfactory receptor 529 |
| chr3_-_76279104 | 0.74 |
ENSRNOT00000007796
|
Olr608
|
olfactory receptor 608 |
| chr9_+_93086012 | 0.72 |
ENSRNOT00000089470
|
Psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr10_-_29196252 | 0.67 |
ENSRNOT00000083910
|
Fabp6
|
fatty acid binding protein 6 |
| chr3_+_76044495 | 0.66 |
ENSRNOT00000049471
|
Olr597
|
olfactory receptor 597 |
| chr4_+_166040503 | 0.65 |
ENSRNOT00000063869
|
Prb3
|
proline-rich protein BstNI subfamily 3 |
| chr13_-_79744939 | 0.65 |
ENSRNOT00000076706
|
Suco
|
SUN domain containing ossification factor |
| chr3_-_76256129 | 0.64 |
ENSRNOT00000072741
|
LOC100912605
|
olfactory receptor 5D14-like |
| chr14_+_9226125 | 0.64 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr13_-_45040593 | 0.64 |
ENSRNOT00000004908
|
Lct
|
lactase |
| chr2_+_46223006 | 0.62 |
ENSRNOT00000073159
|
Olr1265
|
olfactory receptor 1265 |
| chr8_+_36850324 | 0.62 |
ENSRNOT00000060448
|
Pate2
|
prostate and testis expressed 2 |
| chr17_+_45003123 | 0.62 |
ENSRNOT00000084507
|
Olr1658
|
olfactory receptor 1658 |
| chr8_+_42265295 | 0.62 |
ENSRNOT00000074963
|
Olr1220
|
olfactory receptor 1220 |
| chr14_-_21914300 | 0.62 |
ENSRNOT00000087672
|
Csn1s2b
|
casein alpha s2-like B |
| chr3_-_102741818 | 0.55 |
ENSRNOT00000043664
|
Olr766
|
olfactory receptor 766 |
| chr2_+_35298335 | 0.53 |
ENSRNOT00000075359
|
LOC108350226
|
olfactory receptor 147-like |
| chr5_+_90338795 | 0.50 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr1_+_79474809 | 0.48 |
ENSRNOT00000044927
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr8_-_18910837 | 0.44 |
ENSRNOT00000044295
|
Olr1126
|
olfactory receptor 1126 |
| chr4_-_118578815 | 0.43 |
ENSRNOT00000080502
|
Anxa4
|
annexin A4 |
| chr3_+_102529184 | 0.43 |
ENSRNOT00000040444
|
Olr752
|
olfactory receptor 752 |
| chr4_+_121778227 | 0.41 |
ENSRNOT00000084689
|
Vom1r93
|
vomeronasal 1 receptor 93 |
| chr10_-_88019967 | 0.41 |
ENSRNOT00000047580
|
Krt36
|
keratin 36 |
| chr1_+_170796271 | 0.38 |
ENSRNOT00000042820
|
Olr210
|
olfactory receptor 210 |
| chr5_-_50138475 | 0.36 |
ENSRNOT00000011969
|
Slc35a1
|
solute carrier family 35 member A1 |
| chr10_-_87962846 | 0.34 |
ENSRNOT00000018176
|
Krt31
|
keratin 31 |
| chr5_-_38923095 | 0.29 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr1_+_22332090 | 0.26 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr19_-_49448072 | 0.25 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr3_-_72895740 | 0.23 |
ENSRNOT00000012568
|
Fads2l1
|
fatty acid desaturase 2-like 1 |
| chr3_-_73475608 | 0.23 |
ENSRNOT00000049630
|
Olr480
|
olfactory receptor 480 |
| chr11_-_43358275 | 0.22 |
ENSRNOT00000060891
|
Olr1542
|
olfactory receptor 1542 |
| chr2_-_220838905 | 0.14 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr14_-_83412375 | 0.12 |
ENSRNOT00000036848
|
Sfi1
|
SFI1 centrin binding protein |
| chr17_+_69960160 | 0.10 |
ENSRNOT00000023887
|
Ucn3
|
urocortin 3 |
| chr9_+_77320726 | 0.09 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr1_+_60044985 | 0.06 |
ENSRNOT00000079924
|
Vom1r6
|
vomeronasal 1 receptor 6 |
| chr1_+_22364551 | 0.05 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr1_-_236900904 | 0.05 |
ENSRNOT00000066846
|
AABR07006480.1
|
|
| chr1_-_60407295 | 0.03 |
ENSRNOT00000078350
|
Vom1r12
|
vomeronasal 1 receptor 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 2.2 | 6.6 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 1.9 | 5.8 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 1.7 | 16.9 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.7 | 16.6 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 1.6 | 4.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 1.5 | 13.3 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 1.1 | 7.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.0 | 2.9 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.7 | 12.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.7 | 2.7 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.6 | 3.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.5 | 14.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.5 | 22.5 | GO:0042311 | vasodilation(GO:0042311) |
| 0.4 | 17.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.4 | 2.3 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.4 | 4.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 7.0 | GO:0032401 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.3 | 1.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 0.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 2.3 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 1.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 2.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 4.6 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.2 | 6.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 4.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 2.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 7.2 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.6 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.1 | 2.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 2.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 15.4 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 1.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 64.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 4.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.4 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 1.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.6 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.4 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.7 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.9 | 2.7 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.8 | 16.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 40.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 1.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 2.3 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 6.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 0.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 2.0 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 2.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 15.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 7.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 4.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 7.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 3.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 42.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.0 | 2.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 8.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 4.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 16.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 2.2 | 6.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 2.1 | 64.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.7 | 16.9 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 1.6 | 4.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.6 | 4.7 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 1.0 | 7.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.9 | 9.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.8 | 12.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.7 | 4.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.7 | 2.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 2.9 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.4 | 2.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.4 | 1.6 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.3 | 3.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 4.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 22.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 0.6 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.2 | 17.8 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.2 | 2.4 | GO:0016894 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
| 0.2 | 3.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 0.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 5.2 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 1.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.6 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 64.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 22.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 8.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 30.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.1 | 15.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.9 | 12.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 13.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 11.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 2.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 8.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 7.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 4.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |