Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxd8
Z-value: 0.61
Transcription factors associated with Hoxd8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd8
|
ENSRNOG00000042480 | homeobox D8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd8 | rn6_v1_chr3_+_61627383_61627383 | -0.20 | 3.6e-04 | Click! |
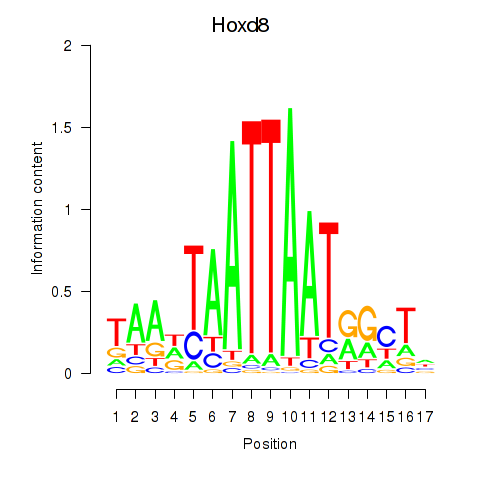
Activity profile of Hoxd8 motif
Sorted Z-values of Hoxd8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_33606124 | 20.78 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr2_+_88217188 | 17.40 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chr3_-_37854561 | 13.89 |
ENSRNOT00000076095
|
Neb
|
nebulin |
| chr4_-_120559078 | 13.78 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr11_-_60547201 | 11.93 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr6_-_140880070 | 11.69 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr18_+_32336102 | 11.32 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr3_-_20419417 | 11.10 |
ENSRNOT00000077772
|
AABR07051733.3
|
|
| chr11_-_60546997 | 10.97 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr11_+_85532526 | 10.77 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr9_+_73418607 | 10.49 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chrX_-_142248369 | 10.02 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr1_+_227670159 | 9.93 |
ENSRNOT00000072077
|
Ms4a6c
|
membrane-spanning 4-domains, subfamily A, member 6C |
| chr5_-_138239306 | 9.62 |
ENSRNOT00000039305
|
Ermap
|
erythroblast membrane-associated protein |
| chr19_-_29968424 | 9.55 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr5_-_12199283 | 9.24 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr13_-_77821312 | 9.22 |
ENSRNOT00000082110
|
AABR07021544.1
|
|
| chr12_-_2245324 | 9.17 |
ENSRNOT00000001332
ENSRNOT00000001333 |
Fcer2
|
Fc fragment of IgE receptor II |
| chr2_-_170301348 | 9.15 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chrX_+_37329779 | 9.08 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr4_-_41212072 | 8.99 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr2_-_200003443 | 8.39 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr6_-_142585188 | 8.36 |
ENSRNOT00000067437
|
AABR07065815.1
|
|
| chr1_+_122981755 | 8.27 |
ENSRNOT00000013468
|
Ndn
|
necdin, MAGE family member |
| chr3_+_63379031 | 8.17 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr11_+_76147205 | 7.75 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr16_+_25773602 | 7.61 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr13_+_92264231 | 7.53 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr7_+_42304534 | 7.41 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr18_+_17043903 | 7.39 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr1_-_128287151 | 7.33 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr12_+_45905371 | 7.23 |
ENSRNOT00000039275
|
Hspb8
|
heat shock protein family B (small) member 8 |
| chr6_+_139405966 | 7.16 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr20_+_34258791 | 7.13 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr3_-_16753987 | 7.07 |
ENSRNOT00000091257
|
AABR07051548.1
|
|
| chr13_+_27465930 | 6.98 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr4_+_93888502 | 6.95 |
ENSRNOT00000090783
|
AABR07060792.1
|
|
| chr4_-_138830117 | 6.83 |
ENSRNOT00000008403
|
Il5ra
|
interleukin 5 receptor subunit alpha |
| chr19_-_37796089 | 6.73 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chrM_+_9451 | 6.50 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr6_-_142353308 | 6.47 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chrX_+_136488691 | 6.46 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr12_+_41486076 | 6.38 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr4_+_29535852 | 6.36 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr2_+_113984646 | 6.04 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr4_-_30380119 | 6.04 |
ENSRNOT00000036460
|
Pon2
|
paraoxonase 2 |
| chr17_+_36691875 | 6.03 |
ENSRNOT00000074314
|
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr13_-_100450209 | 6.02 |
ENSRNOT00000090712
|
Lbr
|
lamin B receptor |
| chr15_+_41643541 | 6.01 |
ENSRNOT00000019646
|
Arl11
|
ADP-ribosylation factor like GTPase 11 |
| chr2_-_231521052 | 5.97 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr19_-_22194740 | 5.93 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr6_+_139428999 | 5.83 |
ENSRNOT00000084482
|
AABR07065693.2
|
|
| chr13_-_102857551 | 5.80 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr8_+_100260049 | 5.69 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr6_-_98666007 | 5.36 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr18_-_85980833 | 5.34 |
ENSRNOT00000058179
|
Socs6
|
suppressor of cytokine signaling 6 |
| chrX_-_76925195 | 5.23 |
ENSRNOT00000087977
|
Atrx
|
ATRX, chromatin remodeler |
| chr2_+_187447501 | 5.09 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr14_-_106234247 | 5.04 |
ENSRNOT00000081245
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr16_+_31734944 | 4.97 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr8_+_133210473 | 4.78 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr17_-_90071408 | 4.78 |
ENSRNOT00000071978
|
Pdss1
|
decaprenyl diphosphate synthase subunit 1 |
| chr4_+_168752133 | 4.77 |
ENSRNOT00000010289
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr9_+_10941613 | 4.75 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chrM_+_9870 | 4.70 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr4_+_163349125 | 4.50 |
ENSRNOT00000084823
|
Klre1
|
killer cell lectin-like receptor, family E, member 1 |
| chr4_-_163402561 | 4.36 |
ENSRNOT00000091890
|
Klrk1
|
killer cell lectin like receptor K1 |
| chr9_+_73378057 | 4.35 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_+_123031307 | 4.28 |
ENSRNOT00000080379
|
Ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr7_-_15073052 | 4.14 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chrM_+_2740 | 3.95 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr17_-_79085076 | 3.94 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr10_-_85725429 | 3.89 |
ENSRNOT00000005471
|
Rpl23
|
ribosomal protein L23 |
| chr16_+_84465656 | 3.89 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr2_+_86996497 | 3.85 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr11_-_70585053 | 3.84 |
ENSRNOT00000086955
|
Zfp148
|
zinc finger protein 148 |
| chr20_+_18594037 | 3.83 |
ENSRNOT00000000753
|
Tfam
|
transcription factor A, mitochondrial |
| chr4_-_107443853 | 3.77 |
ENSRNOT00000066666
|
AABR07061158.1
|
|
| chr17_+_69761118 | 3.67 |
ENSRNOT00000023739
|
Akr1c3
|
aldo-keto reductase family 1, member C3 |
| chr1_-_101095594 | 3.65 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr2_+_239415046 | 3.64 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr2_-_209001895 | 3.61 |
ENSRNOT00000055960
|
RGD1309110
|
similar to Hypothetical protein MGC58999 |
| chr1_-_99632845 | 3.60 |
ENSRNOT00000049123
|
LOC103691107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 pseudogene |
| chr2_-_127710448 | 3.56 |
ENSRNOT00000093733
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr13_-_94859390 | 3.53 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr14_-_41786084 | 3.45 |
ENSRNOT00000059439
|
Grxcr1
|
glutaredoxin and cysteine rich domain containing 1 |
| chr5_+_6373583 | 3.43 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr4_-_148437961 | 3.40 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr9_+_52687868 | 3.31 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr13_-_91735361 | 3.24 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr4_+_88026033 | 3.24 |
ENSRNOT00000074905
|
Vom1r80
|
vomeronasal 1 receptor 80 |
| chr5_+_116420690 | 3.23 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr1_+_65409829 | 3.23 |
ENSRNOT00000046796
|
Vom2r19
|
vomeronasal 2 receptor, 19 |
| chrX_-_40086870 | 3.23 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr1_-_90149991 | 3.23 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr1_+_72692105 | 3.16 |
ENSRNOT00000032439
|
Tmem150b
|
transmembrane protein 150B |
| chr1_+_196095214 | 3.16 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chrX_+_20520034 | 3.12 |
ENSRNOT00000093170
|
FAM120C
|
family with sequence similarity 120C |
| chr4_+_68635581 | 3.10 |
ENSRNOT00000016217
|
Ssbp1
|
single stranded DNA binding protein 1 |
| chrX_-_120521871 | 2.98 |
ENSRNOT00000080863
|
LOC100362376
|
LRRGT00025-like |
| chr2_+_145174876 | 2.94 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr19_-_56983807 | 2.91 |
ENSRNOT00000043529
|
Cox6c-ps1
|
cytochrome c oxidase subunit VIc, pseudogene |
| chr7_+_37101391 | 2.86 |
ENSRNOT00000029764
|
Eea1
|
early endosome antigen 1 |
| chrX_+_54390733 | 2.84 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr13_+_43818010 | 2.83 |
ENSRNOT00000005532
|
LOC100909664
|
centrosomal protein of 170 kDa-like |
| chr15_+_39638510 | 2.81 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr16_-_40025401 | 2.81 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr12_+_47590154 | 2.75 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chrX_+_26440584 | 2.72 |
ENSRNOT00000087293
ENSRNOT00000005328 |
Amelx
|
amelogenin, X-linked |
| chr11_-_82366505 | 2.69 |
ENSRNOT00000041326
|
RGD1559972
|
similar to ribosomal protein L27a |
| chr9_+_67774150 | 2.62 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr18_+_47456327 | 2.60 |
ENSRNOT00000020030
|
Srfbp1
|
serum response factor binding protein 1 |
| chr9_-_11108741 | 2.56 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr16_+_48513432 | 2.56 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr14_-_24080470 | 2.52 |
ENSRNOT00000002745
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr3_-_78545135 | 2.51 |
ENSRNOT00000081552
|
Olr710
|
olfactory receptor 710 |
| chr2_+_86996798 | 2.49 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr14_-_6284145 | 2.48 |
ENSRNOT00000044065
|
Zfp951
|
zinc finger protein 951 |
| chr14_-_6358720 | 2.48 |
ENSRNOT00000048419
|
Zfp951
|
zinc finger protein 951 |
| chrX_+_123770337 | 2.44 |
ENSRNOT00000092554
|
Nkap
|
NFKB activating protein |
| chr1_-_3849080 | 2.43 |
ENSRNOT00000045301
|
RGD1560633
|
similar to ribosomal protein L27a |
| chr6_+_42656616 | 2.40 |
ENSRNOT00000007169
|
Nol10
|
nucleolar protein 10 |
| chr1_+_248132090 | 2.39 |
ENSRNOT00000022056
|
Il33
|
interleukin 33 |
| chr3_-_8955538 | 2.39 |
ENSRNOT00000040570
|
LOC686066
|
similar to 60S ribosomal protein L38 |
| chr9_+_71915421 | 2.36 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr17_-_35130045 | 2.33 |
ENSRNOT00000089878
|
LOC100912478
|
vacuolar ATPase assembly integral membrane protein VMA21-like |
| chr5_-_100977902 | 2.29 |
ENSRNOT00000077429
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr10_+_67810655 | 2.27 |
ENSRNOT00000064285
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr8_-_21453190 | 2.27 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr6_-_38007162 | 2.25 |
ENSRNOT00000044583
|
AABR07063632.1
|
|
| chr3_+_77414999 | 2.24 |
ENSRNOT00000047910
|
Olr659
|
olfactory receptor 659 |
| chr1_+_198409360 | 2.16 |
ENSRNOT00000013691
ENSRNOT00000091295 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr20_-_6257604 | 2.15 |
ENSRNOT00000092489
|
Stk38
|
serine/threonine kinase 38 |
| chrX_-_11750323 | 2.06 |
ENSRNOT00000004303
|
RGD1559951
|
similar to 60S ribosomal protein L37a |
| chr15_+_24078280 | 2.02 |
ENSRNOT00000015511
ENSRNOT00000063807 |
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr3_+_75265525 | 1.97 |
ENSRNOT00000013319
|
Olr554
|
olfactory receptor 554 |
| chr7_-_143167772 | 1.95 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr15_-_93765498 | 1.93 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr3_-_104018861 | 1.87 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr1_-_228263198 | 1.87 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr8_-_61836796 | 1.82 |
ENSRNOT00000064903
ENSRNOT00000084009 |
Commd4
|
COMM domain containing 4 |
| chr11_+_65743892 | 1.80 |
ENSRNOT00000085226
|
AC106292.2
|
|
| chr8_+_5790034 | 1.80 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr5_-_16995521 | 1.76 |
ENSRNOT00000080197
|
Sdr16c6
|
short chain dehydrogenase/reductase family 16C, member 6 |
| chr5_+_165724027 | 1.75 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr4_+_156324922 | 1.74 |
ENSRNOT00000039463
|
Vom2r48
|
vomeronasal 2 receptor, 48 |
| chr8_-_118436894 | 1.73 |
ENSRNOT00000073395
|
LOC100911807
|
ATR-interacting protein-like |
| chr8_+_1459526 | 1.70 |
ENSRNOT00000034503
|
Kbtbd3
|
kelch repeat and BTB domain containing 3 |
| chr8_+_41336340 | 1.68 |
ENSRNOT00000072049
|
Olr1225
|
olfactory receptor 1225 |
| chr13_-_32427177 | 1.63 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr17_+_53965562 | 1.55 |
ENSRNOT00000090387
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr18_-_43945273 | 1.52 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr10_-_87286387 | 1.51 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr14_+_62609484 | 1.50 |
ENSRNOT00000003017
|
Guf1
|
GUF1 homolog, GTPase |
| chr8_+_18795525 | 1.49 |
ENSRNOT00000050430
|
Olr1124
|
olfactory receptor 1124 |
| chr2_-_96509424 | 1.48 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr5_-_166116516 | 1.47 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr3_-_74448279 | 1.47 |
ENSRNOT00000043964
|
Olr527
|
olfactory receptor 527 |
| chr20_-_399933 | 1.42 |
ENSRNOT00000000953
|
Olr1671
|
olfactory receptor 1671 |
| chr2_+_127525285 | 1.40 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr5_+_152680407 | 1.37 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr18_+_35384743 | 1.33 |
ENSRNOT00000076143
ENSRNOT00000074593 |
LOC100911797
|
serine protease inhibitor Kazal-type 5-like |
| chr2_-_61698007 | 1.31 |
ENSRNOT00000049559
|
LOC499541
|
LRRGT00045 |
| chr4_-_66955732 | 1.29 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr11_+_43710705 | 1.22 |
ENSRNOT00000075812
|
Olr1560
|
olfactory receptor 1560 |
| chr12_+_38377034 | 1.22 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr13_-_67206688 | 1.20 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr5_-_162751128 | 1.11 |
ENSRNOT00000068281
|
RGD1559644
|
similar to novel protein similar to esterases |
| chr3_+_163767173 | 1.11 |
ENSRNOT00000010806
|
Ddx27
|
DEAD-box helicase 27 |
| chr1_+_229480184 | 1.11 |
ENSRNOT00000044115
|
Olr387
|
olfactory receptor 387 |
| chr10_+_67810810 | 1.06 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr5_-_16995304 | 1.06 |
ENSRNOT00000061780
|
Sdr16c6
|
short chain dehydrogenase/reductase family 16C, member 6 |
| chrX_-_124252447 | 1.05 |
ENSRNOT00000061546
|
Rhox12
|
reproductive homeobox 12 |
| chr3_-_74815509 | 1.04 |
ENSRNOT00000045173
|
Olr499
|
olfactory receptor 499 |
| chrX_+_74824504 | 1.03 |
ENSRNOT00000048963
|
LOC680353
|
similar to 60S ribosomal protein L38 |
| chr14_+_6221085 | 1.00 |
ENSRNOT00000068215
|
AABR07014259.1
|
|
| chr3_-_77677349 | 0.99 |
ENSRNOT00000084989
|
Olr669
|
olfactory receptor 669 |
| chr3_+_21983841 | 0.99 |
ENSRNOT00000043621
|
RGD1561137
|
similar to 40S ribosomal protein S16 |
| chr3_+_146980923 | 0.96 |
ENSRNOT00000011654
|
Nsfl1c
|
NSFL1 cofactor |
| chr1_+_273854248 | 0.96 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr3_+_31802999 | 0.94 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
| chr3_+_16311578 | 0.89 |
ENSRNOT00000010341
|
Olr409
|
olfactory receptor 409 |
| chr11_+_43143882 | 0.88 |
ENSRNOT00000041445
|
Olr1528
|
olfactory receptor 1528 |
| chr4_+_32541706 | 0.87 |
ENSRNOT00000050827
|
Sdhaf3
|
succinate dehydrogenase complex assembly factor 3 |
| chr17_+_2690062 | 0.85 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chr20_-_32076503 | 0.85 |
ENSRNOT00000000441
|
Supv3l1
|
Suv3 like RNA helicase |
| chr7_-_8060373 | 0.81 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr1_-_164977633 | 0.76 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr10_-_88000423 | 0.75 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr4_-_151428894 | 0.74 |
ENSRNOT00000010556
|
Adipor2
|
adiponectin receptor 2 |
| chr6_+_18880737 | 0.73 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr3_+_48106099 | 0.70 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr10_-_44147035 | 0.70 |
ENSRNOT00000041113
|
Olr1424
|
olfactory receptor 1424 |
| chr3_-_77883067 | 0.69 |
ENSRNOT00000087979
|
Olr677
|
olfactory receptor 677 |
| chr3_-_73934208 | 0.56 |
ENSRNOT00000013038
|
Olr513
|
olfactory receptor 513 |
| chr11_-_83300409 | 0.56 |
ENSRNOT00000029255
|
Vps8
|
VPS8 CORVET complex subunit |
| chr10_-_63274640 | 0.50 |
ENSRNOT00000005129
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr1_-_168930776 | 0.48 |
ENSRNOT00000021468
|
LOC103694854
|
olfactory receptor 52Z1-like |
| chr11_-_45688560 | 0.41 |
ENSRNOT00000081508
|
Olr1536
|
olfactory receptor 1536 |
| chr15_-_65507968 | 0.30 |
ENSRNOT00000046240
|
Calm2
|
calmodulin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 3.0 | 8.9 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 2.3 | 11.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 2.1 | 22.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 1.8 | 7.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 1.7 | 5.2 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.7 | 5.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 1.6 | 4.8 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 1.5 | 7.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.5 | 6.0 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.5 | 7.4 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 1.4 | 10.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.3 | 9.2 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 1.2 | 3.7 | GO:0016488 | response to jasmonic acid(GO:0009753) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of isoprenoid metabolic process(GO:0019747) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.1 | 3.2 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.8 | 3.4 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.7 | 9.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.6 | 2.4 | GO:0061517 | microglial cell activation involved in immune response(GO:0002282) negative regulation of immunoglobulin secretion(GO:0051025) macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.6 | 1.8 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.6 | 3.9 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.5 | 9.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.5 | 1.5 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.5 | 7.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 17.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.4 | 6.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 5.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.4 | 14.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.4 | 1.9 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.4 | 4.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 1.4 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 3.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 3.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.3 | 4.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 6.8 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.3 | 6.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.3 | 1.5 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.3 | 5.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 2.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 0.9 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.3 | 3.6 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.3 | 8.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.3 | 3.4 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.3 | 1.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 1.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 7.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 2.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 3.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 2.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 6.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 1.9 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 | 3.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 4.3 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 2.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 2.6 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.2 | 0.9 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 2.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 4.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 2.9 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 2.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 6.0 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.1 | 6.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 2.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 2.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 4.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 4.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 0.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 5.9 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 1.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 5.3 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 3.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.8 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 2.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 5.8 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 6.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 2.1 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 2.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 3.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 7.1 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 3.2 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 1.7 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.5 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 3.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 3.1 | GO:0019439 | aromatic compound catabolic process(GO:0019439) |
| 0.0 | 5.6 | GO:0006412 | translation(GO:0006412) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.7 | 5.2 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.7 | 15.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.8 | 3.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.8 | 7.5 | GO:0014731 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) cuticular plate(GO:0032437) |
| 0.7 | 5.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.5 | 2.9 | GO:0044308 | axonal spine(GO:0044308) |
| 0.4 | 8.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 3.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 3.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 1.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 6.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 15.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 7.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.3 | 0.9 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.3 | 3.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 1.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 14.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 6.0 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 2.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 5.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 49.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 12.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 13.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 5.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 6.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 8.7 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 2.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 2.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 6.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 20.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 9.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 12.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 7.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 23.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 3.2 | 9.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 3.1 | 9.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.3 | 9.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 2.3 | 9.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.7 | 6.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.6 | 4.8 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 1.4 | 11.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.2 | 3.7 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 1.1 | 3.4 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 1.1 | 7.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.0 | 5.0 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.8 | 5.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.8 | 3.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.7 | 5.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.7 | 3.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.7 | 2.7 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.6 | 3.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.6 | 14.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 11.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 15.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 5.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 9.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 1.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.4 | 4.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 1.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.5 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 2.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 3.6 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 2.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 8.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 10.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 8.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 0.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 0.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 4.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 3.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 2.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 6.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 6.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0031997 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 0.1 | 2.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 4.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 6.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 7.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 7.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 5.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 11.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 4.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.0 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 5.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 2.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 7.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 3.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 9.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 14.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 7.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 8.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 7.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 9.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 6.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 5.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 6.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 25.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 8.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.7 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 9.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.0 | 4.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.9 | 11.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.9 | 11.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.6 | 9.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 12.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.4 | 25.5 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.4 | 7.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.4 | 13.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 7.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 6.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 6.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 1.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 7.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 4.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 6.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.7 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 2.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |