Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxd12
Z-value: 0.46
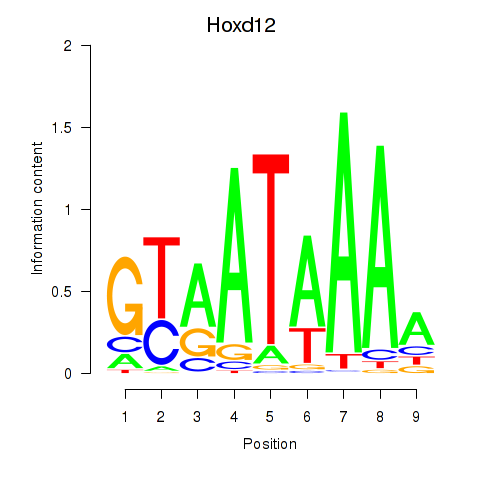
Transcription factors associated with Hoxd12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd12
|
ENSRNOG00000001587 | homeo box D12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd12 | rn6_v1_chr3_+_61597382_61597382 | 0.08 | 1.4e-01 | Click! |
Activity profile of Hoxd12 motif
Sorted Z-values of Hoxd12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_84632817 | 13.16 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr13_+_57243877 | 12.96 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr1_-_93949187 | 12.86 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr11_-_59110562 | 11.72 |
ENSRNOT00000047907
ENSRNOT00000042024 |
Lsamp
|
limbic system-associated membrane protein |
| chr9_+_73529612 | 11.04 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr9_-_60330086 | 10.79 |
ENSRNOT00000092162
ENSRNOT00000093630 |
Dnah7
|
dynein, axonemal, heavy chain 7 |
| chr13_+_77485113 | 9.67 |
ENSRNOT00000080254
|
Tnr
|
tenascin R |
| chr9_+_118586179 | 9.36 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr14_+_12218553 | 9.33 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr13_-_107886476 | 8.78 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr6_+_83421882 | 8.16 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr3_+_48096954 | 7.82 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr18_-_8360684 | 7.64 |
ENSRNOT00000021170
|
Cdh2
|
cadherin 2 |
| chr9_+_95501778 | 7.23 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr3_-_67787990 | 7.17 |
ENSRNOT00000064851
|
Nckap1
|
NCK-associated protein 1 |
| chr10_-_87248572 | 6.88 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr20_+_5008508 | 6.59 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr7_-_93280009 | 6.53 |
ENSRNOT00000064877
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr10_+_77537340 | 6.39 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr7_-_134722215 | 6.22 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chrX_+_53053609 | 5.77 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr2_+_60337667 | 5.67 |
ENSRNOT00000024035
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr8_+_87630916 | 5.57 |
ENSRNOT00000016428
|
Myo6
|
myosin VI |
| chr4_+_99063181 | 5.41 |
ENSRNOT00000008840
|
Fabp1
|
fatty acid binding protein 1 |
| chr15_-_43542939 | 5.03 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr9_+_20048121 | 5.00 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr2_-_250923744 | 4.64 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr2_-_173087648 | 4.55 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr1_+_143036218 | 4.33 |
ENSRNOT00000025797
|
Pde8a
|
phosphodiesterase 8A |
| chr2_+_52431601 | 4.30 |
ENSRNOT00000023554
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr17_+_11683862 | 4.19 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr1_+_148240504 | 4.18 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr8_+_59507990 | 4.06 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chr16_+_22361998 | 4.00 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr16_+_2706428 | 3.99 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chrM_+_7758 | 3.76 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr2_+_260249566 | 3.66 |
ENSRNOT00000089358
|
Slc44a5
|
solute carrier family 44, member 5 |
| chrM_+_5323 | 3.49 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr13_+_91080341 | 3.43 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr10_+_70262361 | 3.42 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr3_+_140024043 | 3.41 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr9_-_30844199 | 3.37 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr3_+_14889510 | 3.16 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr1_-_169344306 | 3.10 |
ENSRNOT00000022852
|
Ubqlnl
|
ubiquilin-like |
| chr14_-_16903242 | 3.03 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr14_-_62595854 | 2.85 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr4_+_158243086 | 2.66 |
ENSRNOT00000032112
|
Ano2
|
anoctamin 2 |
| chr3_+_151126591 | 2.62 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr20_+_41266566 | 2.41 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr3_-_125017377 | 2.41 |
ENSRNOT00000028889
|
Prokr2
|
prokineticin receptor 2 |
| chr4_-_176922424 | 2.39 |
ENSRNOT00000055540
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr8_-_6235967 | 2.29 |
ENSRNOT00000068290
|
LOC654482
|
hypothetical protein LOC654482 |
| chr18_-_26211445 | 2.20 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr2_-_57935334 | 2.19 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chrM_+_7919 | 2.18 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr14_+_39368530 | 2.14 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr1_+_14797766 | 2.03 |
ENSRNOT00000016048
|
Olig3
|
oligodendrocyte transcription factor 3 |
| chr20_+_5049496 | 2.02 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_+_106816152 | 1.93 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr4_-_51199570 | 1.85 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr2_-_173800761 | 1.84 |
ENSRNOT00000039524
|
Wdr49
|
WD repeat domain 49 |
| chr11_+_46108470 | 1.80 |
ENSRNOT00000073467
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr7_-_75288365 | 1.79 |
ENSRNOT00000036904
|
Ankrd46
|
ankyrin repeat domain 46 |
| chr7_+_123168811 | 1.76 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr8_+_107229832 | 1.71 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr20_+_1749716 | 1.70 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr3_+_73352352 | 1.69 |
ENSRNOT00000012814
|
Olr472
|
olfactory receptor 472 |
| chr3_+_93648343 | 1.68 |
ENSRNOT00000091164
|
AABR07053136.1
|
|
| chr14_+_104480662 | 1.66 |
ENSRNOT00000078710
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chrM_+_8599 | 1.54 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr18_+_30880020 | 1.26 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr7_+_140829076 | 1.26 |
ENSRNOT00000086179
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr1_+_204959174 | 1.20 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr19_-_22194740 | 1.15 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr13_+_44615553 | 1.03 |
ENSRNOT00000082439
ENSRNOT00000005289 |
Rab3gap1
|
RAB3 GTPase activating protein catalytic subunit 1 |
| chr18_+_14756684 | 0.99 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_-_17045349 | 0.95 |
ENSRNOT00000086829
|
Fto
|
FTO, alpha-ketoglutarate dependent dioxygenase |
| chr2_+_226563050 | 0.93 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr5_-_50221095 | 0.90 |
ENSRNOT00000012164
|
RGD1563056
|
similar to hypothetical protein |
| chr1_-_238376841 | 0.85 |
ENSRNOT00000076393
|
Tmc1
|
transmembrane channel-like 1 |
| chr5_+_117698764 | 0.79 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr5_+_4373626 | 0.73 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr11_+_46793777 | 0.72 |
ENSRNOT00000002232
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr1_-_129217058 | 0.70 |
ENSRNOT00000086439
|
Pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr16_-_16762240 | 0.69 |
ENSRNOT00000080746
|
AABR07024790.1
|
|
| chr4_+_157726941 | 0.62 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr1_-_131460473 | 0.50 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr7_-_145062956 | 0.35 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr2_+_8732932 | 0.28 |
ENSRNOT00000072849
|
Arrdc3
|
arrestin domain containing 3 |
| chr6_+_52459946 | 0.20 |
ENSRNOT00000073277
ENSRNOT00000013474 |
Atxn7l1
|
ataxin 7-like 1 |
| chr13_+_49005405 | 0.18 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr9_+_19451630 | 0.10 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr3_+_154910626 | 0.09 |
ENSRNOT00000079515
|
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chr1_+_41192824 | 0.03 |
ENSRNOT00000082133
|
Esr1
|
estrogen receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 2.9 | 8.8 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.9 | 5.8 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 1.9 | 5.7 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) |
| 1.8 | 5.4 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 1.6 | 9.7 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.5 | 7.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 1.4 | 7.2 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 1.4 | 12.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.4 | 4.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 1.2 | 9.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.1 | 2.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.9 | 6.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.9 | 19.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.8 | 4.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 3.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.7 | 6.2 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.6 | 10.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.6 | 3.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 | 2.4 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.5 | 13.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.4 | 1.2 | GO:1990168 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.3 | 1.0 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.3 | 2.0 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.3 | 0.9 | GO:0042245 | RNA repair(GO:0042245) |
| 0.3 | 5.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.3 | 3.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 0.3 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.2 | 5.6 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 2.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 4.3 | GO:0009642 | response to light intensity(GO:0009642) cellular response to cholesterol(GO:0071397) |
| 0.2 | 5.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 0.5 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 0.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 4.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 4.3 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 | 1.9 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 3.6 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 7.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 7.2 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 13.0 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 3.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 2.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 2.2 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 2.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 3.0 | GO:0060606 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 7.8 | GO:0007409 | axonogenesis(GO:0007409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 1.9 | 9.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.3 | 7.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.1 | 8.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.1 | 3.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.8 | 7.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 2.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 7.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.5 | 5.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 5.9 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 5.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 5.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 2.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 4.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 0.6 | GO:0070821 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 0.2 | 5.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 13.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 11.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 8.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 15.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 9.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 5.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 4.0 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 5.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 5.1 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 1.8 | 8.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.6 | 12.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.3 | 4.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 1.1 | 5.7 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.9 | 5.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.9 | 4.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.8 | 2.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.7 | 9.4 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.7 | 2.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.6 | 7.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 3.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.5 | 4.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 3.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 13.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.4 | 2.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 2.4 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.4 | 7.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 5.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 7.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 4.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 0.9 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 5.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 4.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 19.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 15.2 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.9 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 10.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 3.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 4.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 5.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 5.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 5.0 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 4.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 11.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 4.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 16.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 5.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.7 | 8.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.6 | 5.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 5.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 3.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 5.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 7.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 7.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 5.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 2.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 5.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 3.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |