Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxd11_Cdx1_Hoxc11
Z-value: 0.77
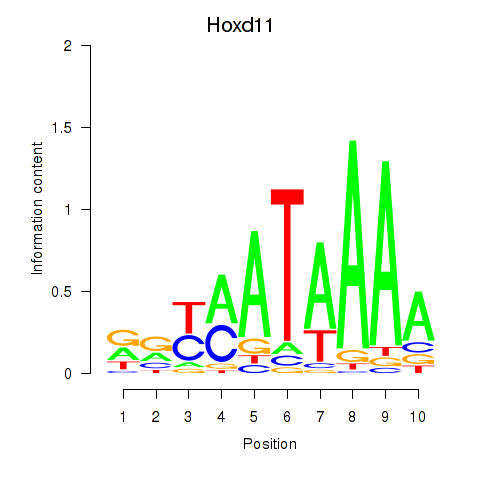
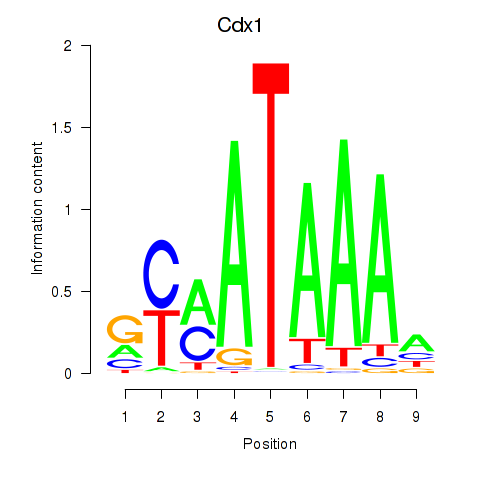
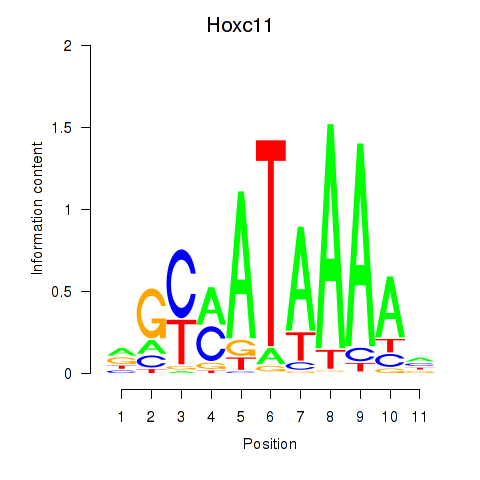
Transcription factors associated with Hoxd11_Cdx1_Hoxc11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd11
|
ENSRNOG00000051472 | homeobox D11 |
|
Cdx1
|
ENSRNOG00000060334 | caudal type homeo box 1 |
|
Hoxc11
|
ENSRNOG00000016141 | homeobox C11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd11 | rn6_v1_chr3_+_61604672_61604672 | 0.54 | 2.0e-25 | Click! |
| Hoxc11 | rn6_v1_chr7_+_144565392_144565392 | 0.38 | 2.8e-12 | Click! |
| Cdx1 | rn6_v1_chr18_-_56355490_56355490 | -0.04 | 4.8e-01 | Click! |
Activity profile of Hoxd11_Cdx1_Hoxc11 motif
Sorted Z-values of Hoxd11_Cdx1_Hoxc11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_20048121 | 52.47 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr4_-_51199570 | 43.11 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr16_+_2706428 | 33.82 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr1_-_207811008 | 28.42 |
ENSRNOT00000080506
|
Clrn3
|
clarin 3 |
| chr14_-_20920286 | 23.26 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr4_+_99063181 | 22.73 |
ENSRNOT00000008840
|
Fabp1
|
fatty acid binding protein 1 |
| chr4_-_173640684 | 22.30 |
ENSRNOT00000010728
|
Rergl
|
RERG like |
| chr2_-_100249811 | 20.37 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr3_+_159902441 | 20.36 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr10_-_88060561 | 20.08 |
ENSRNOT00000019133
|
Krt19
|
keratin 19 |
| chr20_+_3677474 | 19.62 |
ENSRNOT00000047663
|
Mt1m
|
metallothionein 1M |
| chr9_-_60330086 | 18.53 |
ENSRNOT00000092162
ENSRNOT00000093630 |
Dnah7
|
dynein, axonemal, heavy chain 7 |
| chr8_+_67753279 | 17.76 |
ENSRNOT00000009716
|
Calml4
|
calmodulin-like 4 |
| chr1_+_213511874 | 16.51 |
ENSRNOT00000078080
ENSRNOT00000016883 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr5_-_147784311 | 16.48 |
ENSRNOT00000074172
|
Fam167b
|
family with sequence similarity 167, member B |
| chr14_+_12218553 | 16.36 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr5_+_117698764 | 15.03 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr9_-_52238564 | 14.51 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr3_+_151126591 | 13.24 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr1_+_41192824 | 12.92 |
ENSRNOT00000082133
|
Esr1
|
estrogen receptor 1 |
| chr20_-_14020007 | 12.55 |
ENSRNOT00000093521
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_-_6235967 | 12.27 |
ENSRNOT00000068290
|
LOC654482
|
hypothetical protein LOC654482 |
| chr18_-_24823837 | 10.98 |
ENSRNOT00000021405
ENSRNOT00000090390 |
Myo7b
|
myosin VIIb |
| chr19_-_22194740 | 10.91 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr2_-_154418629 | 9.99 |
ENSRNOT00000076274
ENSRNOT00000076165 |
Plch1
|
phospholipase C, eta 1 |
| chr2_+_60920257 | 9.70 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr13_+_57243877 | 9.38 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr2_+_8732932 | 9.21 |
ENSRNOT00000072849
|
Arrdc3
|
arrestin domain containing 3 |
| chr17_-_18590536 | 8.98 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_167141875 | 8.90 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr4_-_82289672 | 8.87 |
ENSRNOT00000036394
|
Hoxa10
|
homeobox A10 |
| chr2_-_204254699 | 8.75 |
ENSRNOT00000021487
|
Mab21l3
|
mab-21 like 3 |
| chr13_-_50514151 | 8.60 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr20_+_41266566 | 8.44 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr9_+_81689802 | 8.42 |
ENSRNOT00000021432
|
Vil1
|
villin 1 |
| chr4_-_178441547 | 8.19 |
ENSRNOT00000087434
ENSRNOT00000055542 |
Sox5
|
SRY box 5 |
| chr19_-_37916813 | 7.92 |
ENSRNOT00000026585
|
Lcat
|
lecithin cholesterol acyltransferase |
| chrX_+_112769645 | 7.88 |
ENSRNOT00000064478
|
Col4a5
|
collagen type IV alpha 5 chain |
| chr13_-_111972603 | 7.81 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr7_+_116090189 | 7.79 |
ENSRNOT00000081571
|
Psca
|
prostate stem cell antigen |
| chr5_+_162808646 | 7.64 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr18_-_8360684 | 7.42 |
ENSRNOT00000021170
|
Cdh2
|
cadherin 2 |
| chr8_+_49621568 | 7.09 |
ENSRNOT00000021949
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr4_+_155313671 | 6.88 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chr12_-_40590361 | 6.84 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr8_+_80965255 | 6.76 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr7_+_38742051 | 6.76 |
ENSRNOT00000006070
|
Dcn
|
decorin |
| chr11_+_46108470 | 6.71 |
ENSRNOT00000073467
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr6_-_42002819 | 6.50 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chrX_-_139079336 | 6.34 |
ENSRNOT00000047605
|
Usp26
|
ubiquitin specific peptidase 26 |
| chr16_+_54291251 | 6.24 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr10_+_77537340 | 6.24 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chrX_+_110789269 | 6.02 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr7_+_76092503 | 6.01 |
ENSRNOT00000079692
|
Grhl2
|
grainyhead-like transcription factor 2 |
| chr19_+_9895121 | 6.00 |
ENSRNOT00000033953
|
Prss54
|
protease, serine, 54 |
| chr3_+_93648343 | 5.87 |
ENSRNOT00000091164
|
AABR07053136.1
|
|
| chr4_+_167017816 | 5.59 |
ENSRNOT00000055734
|
Tas2r145
|
taste receptor, type 2, member 145 |
| chr2_+_200572502 | 5.57 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chrM_+_3904 | 5.51 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_-_89369960 | 5.06 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr2_-_88553086 | 4.91 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr1_-_238376841 | 4.84 |
ENSRNOT00000076393
|
Tmc1
|
transmembrane channel-like 1 |
| chr6_+_73358112 | 4.69 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr5_+_167142182 | 4.69 |
ENSRNOT00000024054
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr9_+_52023295 | 4.68 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr8_-_63750531 | 4.67 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr3_+_140024043 | 4.67 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr4_-_28702559 | 4.52 |
ENSRNOT00000013910
ENSRNOT00000013937 |
Calcr
|
calcitonin receptor |
| chr8_-_109560747 | 4.51 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr20_+_5008508 | 4.44 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr11_+_46793777 | 4.34 |
ENSRNOT00000002232
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr14_-_66978499 | 4.29 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr7_+_130042947 | 4.13 |
ENSRNOT00000089707
|
Panx2
|
pannexin 2 |
| chr1_-_126211439 | 4.04 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr3_-_78112770 | 3.96 |
ENSRNOT00000008396
|
Olr694
|
olfactory receptor 694 |
| chr1_-_169344306 | 3.82 |
ENSRNOT00000022852
|
Ubqlnl
|
ubiquilin-like |
| chr8_+_117906014 | 3.77 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr16_+_22361998 | 3.74 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr2_-_154418920 | 3.72 |
ENSRNOT00000076326
|
Plch1
|
phospholipase C, eta 1 |
| chr1_+_78546012 | 3.71 |
ENSRNOT00000058172
|
Gng5
|
G protein subunit gamma 5 |
| chr18_-_26211445 | 3.69 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr8_+_119030875 | 3.64 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr17_-_42127678 | 3.49 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr7_-_134722215 | 3.48 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr7_-_118332577 | 3.44 |
ENSRNOT00000090370
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr6_-_22138286 | 3.37 |
ENSRNOT00000007607
|
Yipf4
|
Yip1 domain family, member 4 |
| chr14_+_39368530 | 3.30 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr15_+_50891127 | 3.26 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr10_+_84119884 | 3.21 |
ENSRNOT00000009951
|
Hoxb9
|
homeo box B9 |
| chr4_-_157486844 | 3.16 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr1_+_193424812 | 3.11 |
ENSRNOT00000019939
|
Aqp8
|
aquaporin 8 |
| chrX_+_33884499 | 3.11 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr5_+_172488708 | 3.08 |
ENSRNOT00000019930
|
Morn1
|
MORN repeat containing 1 |
| chrM_+_8599 | 3.05 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr8_+_42613601 | 2.98 |
ENSRNOT00000040297
|
Olr1264
|
olfactory receptor 1264 |
| chr15_+_17775692 | 2.96 |
ENSRNOT00000061169
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr5_+_71742911 | 2.93 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr7_-_69982592 | 2.87 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr19_+_53044379 | 2.86 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr9_+_73529612 | 2.85 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chrX_+_104734082 | 2.77 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chrM_+_7758 | 2.75 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr2_-_79078258 | 2.71 |
ENSRNOT00000031170
|
Fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr7_+_64768742 | 2.70 |
ENSRNOT00000005545
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_250805445 | 2.69 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr17_-_22143324 | 2.64 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chrM_+_7919 | 2.59 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr16_+_50152008 | 2.53 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr6_+_76041669 | 2.49 |
ENSRNOT00000039829
|
Fam177a1
|
family with sequence similarity 177, member A1 |
| chr7_+_34533543 | 2.43 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr5_-_50221095 | 2.35 |
ENSRNOT00000012164
|
RGD1563056
|
similar to hypothetical protein |
| chr13_+_91080341 | 2.35 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr2_+_54466280 | 2.32 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr5_-_6186329 | 2.19 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr20_+_5049496 | 2.17 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr4_-_84131030 | 2.15 |
ENSRNOT00000012196
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr6_-_8344574 | 2.14 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr10_+_71546977 | 2.14 |
ENSRNOT00000041140
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr9_+_118586179 | 2.13 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr16_-_16762240 | 2.10 |
ENSRNOT00000080746
|
AABR07024790.1
|
|
| chr4_-_167068838 | 2.06 |
ENSRNOT00000029713
|
Tas2r125
|
taste receptor, type 2, member 125 |
| chr13_-_37697569 | 2.04 |
ENSRNOT00000045263
|
LOC680288
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr13_+_26463404 | 2.01 |
ENSRNOT00000061779
|
LOC103690041
|
serpin B12-like |
| chrM_+_2740 | 1.99 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr2_-_231409988 | 1.98 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr14_+_104480662 | 1.97 |
ENSRNOT00000078710
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chr3_+_54253949 | 1.97 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr17_+_32973695 | 1.90 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr7_+_142575672 | 1.89 |
ENSRNOT00000080923
ENSRNOT00000008160 |
Scn8a
|
sodium voltage-gated channel alpha subunit 8 |
| chr16_+_74785281 | 1.85 |
ENSRNOT00000064316
ENSRNOT00000017106 ENSRNOT00000090646 |
Nek3
|
NIMA-related kinase 3 |
| chr6_+_94835845 | 1.83 |
ENSRNOT00000006321
|
Jkamp
|
JNK1/MAPK8-associated membrane protein |
| chr1_+_125124743 | 1.82 |
ENSRNOT00000021530
|
Mtmr10
|
myotubularin related protein 10 |
| chr5_-_113880911 | 1.77 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr4_-_176679815 | 1.73 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr1_-_198652674 | 1.70 |
ENSRNOT00000023706
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr7_+_58814805 | 1.67 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr1_+_44311513 | 1.62 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr2_+_188745503 | 1.62 |
ENSRNOT00000056652
|
Shc1
|
SHC adaptor protein 1 |
| chr3_-_52510507 | 1.57 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr3_-_72219246 | 1.55 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr2_-_231409496 | 1.54 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr18_+_35136132 | 1.53 |
ENSRNOT00000078731
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr17_+_31236015 | 1.49 |
ENSRNOT00000023149
|
Slc22a23
|
solute carrier family 22, member 23 |
| chr14_+_9555264 | 1.46 |
ENSRNOT00000002928
|
Nkx6-1
|
NK6 homeobox 1 |
| chr2_+_22910236 | 1.46 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr12_+_9464026 | 1.46 |
ENSRNOT00000048188
|
Cdx2
|
caudal type homeo box 2 |
| chr10_+_78416946 | 1.40 |
ENSRNOT00000042211
|
AABR07030263.1
|
|
| chr9_-_85617954 | 1.38 |
ENSRNOT00000077331
|
Serpine2
|
serpin family E member 2 |
| chr8_-_23000455 | 1.34 |
ENSRNOT00000091027
ENSRNOT00000017493 |
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr4_-_120157477 | 1.34 |
ENSRNOT00000046568
|
AABR07061382.1
|
|
| chr8_+_118066988 | 1.32 |
ENSRNOT00000056161
|
Map4
|
microtubule-associated protein 4 |
| chr5_-_17399801 | 1.29 |
ENSRNOT00000011900
|
RGD1563405
|
similar to protein tyrosine phosphatase 4a2 |
| chr18_+_29999290 | 1.28 |
ENSRNOT00000027372
|
Pcdha4
|
protocadherin alpha 4 |
| chr11_-_89260297 | 1.27 |
ENSRNOT00000057502
|
Spidr
|
scaffolding protein involved in DNA repair |
| chr1_-_168611670 | 1.25 |
ENSRNOT00000021273
|
Olr106
|
olfactory receptor 106 |
| chr1_+_31409579 | 1.24 |
ENSRNOT00000016989
|
RGD1311933
|
similar to RIKEN cDNA 2310057J18 |
| chr2_+_52431601 | 1.23 |
ENSRNOT00000023554
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr2_+_193778042 | 1.20 |
ENSRNOT00000090212
|
Tchh
|
trichohyalin |
| chr17_-_14627937 | 1.19 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr3_+_43255567 | 1.17 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr1_+_14797766 | 1.12 |
ENSRNOT00000016048
|
Olig3
|
oligodendrocyte transcription factor 3 |
| chr20_+_3364814 | 1.10 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr13_+_95887708 | 1.09 |
ENSRNOT00000029081
|
LOC689766
|
hypothetical protein LOC689766 |
| chr7_+_64769089 | 1.04 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr1_+_230811800 | 0.98 |
ENSRNOT00000041509
|
Olr384
|
olfactory receptor 384 |
| chr8_+_130350510 | 0.98 |
ENSRNOT00000073376
|
Ss18l2
|
SS18 like 2 |
| chr3_+_48096954 | 0.98 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr20_+_29558689 | 0.95 |
ENSRNOT00000085229
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr8_-_60570058 | 0.95 |
ENSRNOT00000009169
|
Scaper
|
S-phase cyclin A-associated protein in the ER |
| chr13_+_71305548 | 0.94 |
ENSRNOT00000074864
|
LOC684709
|
similar to putative membrane protein Re9 |
| chr2_+_243577082 | 0.93 |
ENSRNOT00000016556
|
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr18_+_17403407 | 0.91 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr7_-_13404858 | 0.91 |
ENSRNOT00000060456
|
Olr1075
|
olfactory receptor 1075 |
| chr10_-_63274640 | 0.90 |
ENSRNOT00000005129
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr20_-_5064469 | 0.87 |
ENSRNOT00000001120
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr20_-_1099336 | 0.87 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chr3_-_78790691 | 0.87 |
ENSRNOT00000008750
|
Olr724
|
olfactory receptor 724 |
| chr6_+_37144787 | 0.85 |
ENSRNOT00000075503
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr9_+_16737642 | 0.80 |
ENSRNOT00000061430
|
Srf
|
serum response factor |
| chr1_+_99616447 | 0.79 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr1_+_80880856 | 0.78 |
ENSRNOT00000026060
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr7_-_145062956 | 0.73 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr9_-_81879211 | 0.73 |
ENSRNOT00000085140
|
Rnf25
|
ring finger protein 25 |
| chrX_+_6273733 | 0.72 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr5_+_128881062 | 0.69 |
ENSRNOT00000057216
|
Calr4
|
calreticulin 4 |
| chr10_+_84031955 | 0.69 |
ENSRNOT00000009934
|
Hoxb13
|
homeo box B13 |
| chr11_+_28780446 | 0.68 |
ENSRNOT00000072546
|
Krtap15-1
|
keratin associated protein 15-1 |
| chr6_-_103949275 | 0.63 |
ENSRNOT00000047825
|
AABR07065012.1
|
|
| chr1_-_172215730 | 0.63 |
ENSRNOT00000055175
|
Olr244
|
olfactory receptor 244 |
| chr13_+_90533365 | 0.57 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr9_+_66888393 | 0.52 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr7_+_140829076 | 0.50 |
ENSRNOT00000086179
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr8_+_106816152 | 0.50 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr1_+_150492239 | 0.49 |
ENSRNOT00000047771
ENSRNOT00000048404 |
AABR07004684.1
|
|
| chr18_-_6587080 | 0.49 |
ENSRNOT00000040815
|
LOC103694404
|
60S ribosomal protein L39 |
| chr11_+_43486904 | 0.48 |
ENSRNOT00000083800
|
Olr1549
|
olfactory receptor 1549 |
| chr2_+_112868707 | 0.48 |
ENSRNOT00000017805
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr2_+_166856784 | 0.46 |
ENSRNOT00000035765
|
Otol1
|
otolin 1 |
| chr10_+_45322248 | 0.43 |
ENSRNOT00000047268
|
Trim11
|
tripartite motif-containing 11 |
| chrX_-_23144324 | 0.43 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd11_Cdx1_Hoxc11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.7 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 5.5 | 16.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 5.1 | 20.4 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 3.6 | 14.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 3.3 | 16.5 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 3.2 | 12.9 | GO:1990375 | baculum development(GO:1990375) |
| 3.1 | 12.5 | GO:0031179 | peptide modification(GO:0031179) |
| 3.0 | 15.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 2.7 | 13.6 | GO:0015755 | fructose transport(GO:0015755) |
| 2.7 | 43.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 2.6 | 7.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.3 | 9.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 2.2 | 11.0 | GO:1904970 | brush border assembly(GO:1904970) |
| 2.1 | 8.4 | GO:1902896 | regulation of microvillus length(GO:0032532) terminal web assembly(GO:1902896) |
| 1.7 | 3.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.7 | 8.6 | GO:0002003 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 1.5 | 4.5 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 1.5 | 6.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.5 | 7.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 1.4 | 4.3 | GO:0090024 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.4 | 9.7 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 1.4 | 8.2 | GO:2000741 | regulation of timing of neuron differentiation(GO:0060164) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.3 | 24.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 1.3 | 5.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 1.2 | 3.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 4.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.1 | 18.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 1.0 | 4.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 1.0 | 6.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.9 | 6.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.9 | 3.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.8 | 7.9 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.8 | 2.3 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.7 | 2.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.7 | 2.9 | GO:0072011 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) glomerular mesangial cell development(GO:0072144) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.7 | 2.6 | GO:0014826 | vein smooth muscle contraction(GO:0014826) response to prostaglandin F(GO:0034696) rhythmic excitation(GO:0043179) positive regulation of sarcomere organization(GO:0060298) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.6 | 20.1 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.6 | 3.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.6 | 3.5 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.6 | 4.0 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.5 | 2.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.5 | 3.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 2.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.5 | 7.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.5 | 3.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.5 | 4.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.4 | 3.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.4 | 1.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 0.8 | GO:0060947 | cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 0.4 | 2.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 2.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 1.4 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.3 | 8.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 1.5 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.3 | 2.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 1.5 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 3.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 7.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 3.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 5.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 2.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 5.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 1.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 0.7 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 1.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.2 | 6.0 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 1.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 1.9 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 6.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 2.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 5.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 7.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 4.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.2 | 5.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.3 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.2 | 3.1 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 3.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 6.9 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 8.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 4.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 2.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 4.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 3.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 11.3 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 1.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.6 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 16.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.8 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.4 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.9 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.1 | 1.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.5 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 3.7 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 3.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 8.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.9 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 3.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.5 | GO:0048644 | muscle organ morphogenesis(GO:0048644) |
| 0.0 | 7.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.6 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 2.0 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.2 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.0 | 0.5 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 1.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 18.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 3.6 | 14.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.9 | 20.1 | GO:1990357 | terminal web(GO:1990357) |
| 2.8 | 27.8 | GO:0045179 | apical cortex(GO:0045179) |
| 1.5 | 13.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.4 | 10.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 1.3 | 12.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.2 | 3.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.1 | 19.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 1.0 | 3.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.6 | 6.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.6 | 7.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.4 | 1.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.4 | 2.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 11.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 5.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 4.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 5.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 2.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 4.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 14.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 1.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 5.1 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 3.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 9.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 10.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 3.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 10.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 1.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 4.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 16.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 132.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 23.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 3.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 24.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 7.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 7.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 32.8 | GO:0098796 | membrane protein complex(GO:0098796) |
| 0.1 | 19.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 4.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.8 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.8 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.2 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 4.2 | 33.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 3.9 | 23.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 3.8 | 22.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 3.3 | 16.4 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 2.7 | 43.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 2.6 | 7.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.6 | 12.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 2.1 | 12.5 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 1.9 | 7.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.7 | 20.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 1.7 | 15.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 1.6 | 7.9 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.5 | 13.6 | GO:0070061 | fructose binding(GO:0070061) |
| 1.4 | 10.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.2 | 3.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 1.1 | 4.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 1.1 | 4.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 1.0 | 4.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.9 | 2.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.8 | 16.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.7 | 2.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.6 | 4.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.6 | 7.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 2.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 4.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 2.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.5 | 2.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 6.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.5 | 9.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 3.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 4.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 52.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 27.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 2.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 5.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 2.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.4 | 2.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 1.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.4 | 9.2 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.4 | 16.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 1.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.4 | 7.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 2.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.3 | 8.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 9.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 7.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 18.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 1.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 36.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 1.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 2.1 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.2 | 3.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 3.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 3.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 8.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 6.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 3.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 5.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.8 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 3.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 5.9 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 5.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 0.4 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 17.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 5.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 5.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 3.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.5 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 33.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.6 | 27.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.4 | 21.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 26.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 15.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 6.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 54.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 3.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 7.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 4.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 6.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 9.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 3.1 | 43.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.6 | 42.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.5 | 16.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.9 | 13.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.6 | 10.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.6 | 27.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.6 | 6.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 7.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 12.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 12.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 4.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 13.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 3.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 7.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 12.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 2.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 3.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 3.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.2 | 16.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 13.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 12.7 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 2.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 2.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 3.0 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |