Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
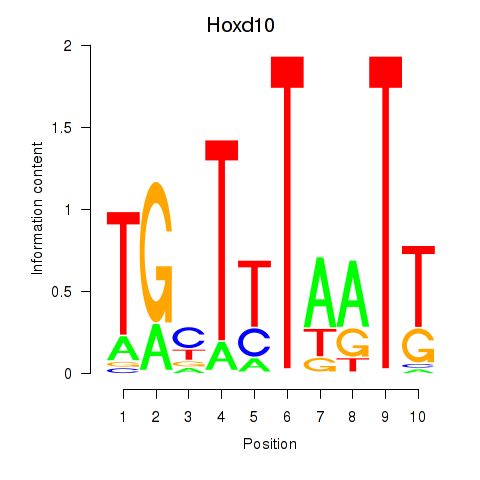
Results for Hoxd10
Z-value: 0.23
Transcription factors associated with Hoxd10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd10
|
ENSRNOG00000001581 | homeo box D10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd10 | rn6_v1_chr3_+_61613774_61613774 | -0.18 | 1.1e-03 | Click! |
Activity profile of Hoxd10 motif
Sorted Z-values of Hoxd10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_31138675 | 6.12 |
ENSRNOT00000004456
|
Fancb
|
Fanconi anemia, complementation group B |
| chr15_-_77736892 | 4.00 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr6_-_95061174 | 3.31 |
ENSRNOT00000072262
|
Rtn1
|
reticulon 1 |
| chr18_+_48853418 | 3.24 |
ENSRNOT00000022792
|
Csnk1g3
|
casein kinase 1, gamma 3 |
| chr4_-_164453171 | 2.97 |
ENSRNOT00000077539
ENSRNOT00000083610 ENSRNOT00000079975 |
Ly49s6
|
Ly49 stimulatory receptor 6 |
| chr14_-_45859908 | 2.70 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr19_+_25043680 | 2.60 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr11_-_11214141 | 2.54 |
ENSRNOT00000065748
|
Robo2
|
roundabout guidance receptor 2 |
| chrX_-_76708878 | 2.41 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr13_-_99531959 | 2.35 |
ENSRNOT00000005059
|
Wdr26
|
WD repeat domain 26 |
| chr11_-_11213821 | 2.25 |
ENSRNOT00000093706
|
Robo2
|
roundabout guidance receptor 2 |
| chrX_-_138435391 | 2.25 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr9_+_73334618 | 2.03 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr9_+_119542328 | 1.99 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr16_-_48692476 | 1.82 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr8_-_78397123 | 1.70 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr13_+_82496022 | 1.68 |
ENSRNOT00000080759
|
F5
|
coagulation factor V |
| chr1_+_252409268 | 1.16 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr7_+_38858062 | 1.10 |
ENSRNOT00000006234
|
Kera
|
keratocan |
| chr1_+_57692836 | 0.99 |
ENSRNOT00000083968
ENSRNOT00000019358 |
Chd1
|
chromodomain helicase DNA binding protein 1 |
| chr12_+_7081895 | 0.90 |
ENSRNOT00000047163
|
Hmgb1
|
high mobility group box 1 |
| chr19_+_32308236 | 0.87 |
ENSRNOT00000015508
|
Mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr4_+_24222500 | 0.79 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr10_-_65502936 | 0.76 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr2_+_243425007 | 0.74 |
ENSRNOT00000082894
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr7_+_97778260 | 0.54 |
ENSRNOT00000064264
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr14_-_11546314 | 0.39 |
ENSRNOT00000090334
|
AABR07014355.1
|
|
| chr8_+_18785118 | 0.38 |
ENSRNOT00000090997
|
LOC686748
|
similar to olfactory receptor 829 |
| chr1_+_244630377 | 0.23 |
ENSRNOT00000016740
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_75437833 | 0.07 |
ENSRNOT00000042246
|
Olr561
|
olfactory receptor 561 |
| chr11_+_80319177 | 0.07 |
ENSRNOT00000036777
|
Rtp2
|
receptor (chemosensory) transporter protein 2 |
| chr1_-_169845487 | 0.07 |
ENSRNOT00000039989
|
Olr179
|
olfactory receptor 179 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 1.0 | 4.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.8 | 2.4 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.3 | 0.9 | GO:0034165 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) regulation of mismatch repair(GO:0032423) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) T-helper 1 cell activation(GO:0035711) positive regulation of glycogen catabolic process(GO:0045819) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.2 | 1.7 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 2.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.8 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.1 | 1.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 2.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.0 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 2.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 2.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 3.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.7 | GO:0006006 | glucose metabolic process(GO:0006006) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.4 | 6.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 2.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 4.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 4.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 2.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 4.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 0.8 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.2 | 0.9 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 2.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 3.2 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 1.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |