Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxd1
Z-value: 0.59
Transcription factors associated with Hoxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd1
|
ENSRNOG00000001572 | homeo box D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd1 | rn6_v1_chr3_+_61685619_61685619 | 0.07 | 2.1e-01 | Click! |
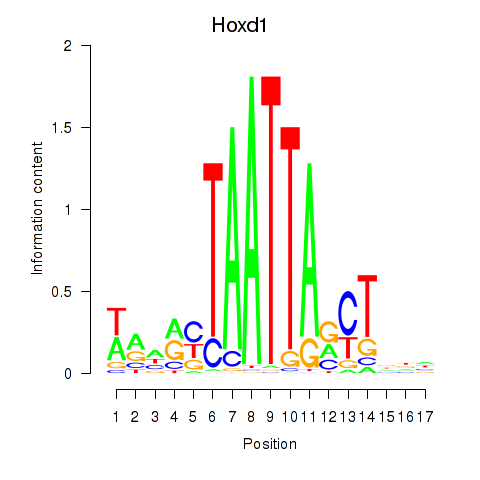
Activity profile of Hoxd1 motif
Sorted Z-values of Hoxd1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_227457629 | 18.60 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr6_+_2216623 | 15.03 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr9_-_4978892 | 14.92 |
ENSRNOT00000015189
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr8_-_77398156 | 13.59 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr14_-_21748356 | 12.79 |
ENSRNOT00000002670
|
Cabs1
|
calcium binding protein, spermatid associated 1 |
| chr3_+_159368273 | 12.47 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr14_+_22517774 | 12.02 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr16_-_24951612 | 11.73 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr7_-_71139267 | 11.39 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr5_+_28485619 | 10.08 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr2_+_195651930 | 9.71 |
ENSRNOT00000028299
|
Tdrkh
|
tudor and KH domain containing |
| chr6_+_56846789 | 9.11 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr1_+_253221812 | 8.83 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr12_-_2438817 | 8.71 |
ENSRNOT00000037059
|
Ccl25
|
C-C motif chemokine ligand 25 |
| chr10_+_103206014 | 8.66 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr14_+_22375955 | 8.44 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_-_233145924 | 8.41 |
ENSRNOT00000018763
ENSRNOT00000082442 |
Psat1
|
phosphoserine aminotransferase 1 |
| chr1_+_140998240 | 8.26 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr10_-_110274768 | 7.89 |
ENSRNOT00000054931
|
Sectm1a
|
secreted and transmembrane 1A |
| chr2_-_132301073 | 7.76 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr1_+_190200324 | 7.64 |
ENSRNOT00000001933
|
Abca16
|
ATP-binding cassette, subfamily A (ABC1), member 16 |
| chr6_-_7058314 | 7.54 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr3_-_14229067 | 7.41 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr1_+_201337416 | 7.30 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr8_-_12993651 | 7.16 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr2_+_54466280 | 7.14 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr4_+_70977556 | 7.12 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr10_-_78219793 | 6.98 |
ENSRNOT00000003369
|
Tom1l1
|
target of myb1 like 1 membrane trafficking protein |
| chr8_-_113937097 | 6.96 |
ENSRNOT00000046816
|
Nek11
|
NIMA-related kinase 11 |
| chr15_+_33885106 | 6.95 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr4_+_52235811 | 6.83 |
ENSRNOT00000060478
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr2_-_216382244 | 6.80 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr16_+_71889235 | 6.74 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr9_+_95161157 | 6.69 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr7_-_54855557 | 6.58 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chrX_+_144994139 | 6.43 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr9_+_60900213 | 6.31 |
ENSRNOT00000017859
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr5_-_147412705 | 6.26 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr2_-_181900856 | 6.15 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr4_-_168656673 | 6.07 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr12_-_19167015 | 6.04 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr1_+_101603222 | 6.02 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr5_-_59165160 | 5.96 |
ENSRNOT00000029035
|
Fam221b
|
family with sequence similarity 221, member B |
| chr7_-_15483834 | 5.84 |
ENSRNOT00000057608
|
Morc2b
|
microrchidia 2B |
| chr13_-_76049363 | 5.83 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr18_+_4084228 | 5.83 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr2_-_183128932 | 5.82 |
ENSRNOT00000031179
|
Mnd1
|
meiotic nuclear divisions 1 |
| chrX_+_14019961 | 5.78 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr10_-_88670430 | 5.78 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr3_-_21904133 | 5.75 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr20_-_5166448 | 5.73 |
ENSRNOT00000076331
|
Aif1
|
allograft inflammatory factor 1 |
| chr5_-_12526962 | 5.63 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr19_-_42180362 | 5.59 |
ENSRNOT00000089515
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr18_+_81694808 | 5.57 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr2_+_66940057 | 5.56 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr18_+_63203063 | 5.47 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr4_-_80395502 | 5.34 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr8_+_107495366 | 5.32 |
ENSRNOT00000056590
|
Cep70
|
centrosomal protein 70 |
| chr8_+_128027958 | 5.26 |
ENSRNOT00000045049
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr7_-_55604403 | 5.25 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr2_-_216348194 | 5.22 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr14_+_23480462 | 5.22 |
ENSRNOT00000002755
|
Gnrhr
|
gonadotropin releasing hormone receptor |
| chr1_-_73753128 | 5.19 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr9_-_95362014 | 5.14 |
ENSRNOT00000051065
|
Hjurp
|
Holliday junction recognition protein |
| chr9_+_20241062 | 5.13 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr4_-_157304653 | 5.11 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr1_+_88113445 | 5.05 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr17_-_43537293 | 4.85 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr16_+_73615949 | 4.72 |
ENSRNOT00000024392
|
Gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chrX_-_98095663 | 4.70 |
ENSRNOT00000004491
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr1_-_14117021 | 4.68 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr6_-_108660063 | 4.55 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr4_-_161907767 | 4.54 |
ENSRNOT00000009557
|
A2ml1
|
alpha-2-macroglobulin-like 1 |
| chr8_+_41350182 | 4.54 |
ENSRNOT00000072823
|
LOC100911758
|
olfactory receptor 143-like |
| chr11_-_61234944 | 4.54 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr12_-_46493203 | 4.50 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr1_-_81550598 | 4.40 |
ENSRNOT00000051671
|
RGD1564380
|
similar to BC049730 protein |
| chr9_+_73378057 | 4.38 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_+_129885826 | 4.36 |
ENSRNOT00000009743
|
Slx4ip
|
SLX4 interacting protein |
| chr13_-_50514151 | 4.34 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr4_+_145238947 | 4.32 |
ENSRNOT00000067396
ENSRNOT00000091934 |
Cpne9
|
copine family member 9 |
| chr10_-_87564327 | 4.28 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr10_+_82032656 | 4.27 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr7_-_28711761 | 4.22 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr9_+_35289443 | 4.21 |
ENSRNOT00000037765
ENSRNOT00000060503 |
Spata31e1
|
SPATA31 subfamily E, member 1 |
| chr10_-_44746549 | 4.20 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chrX_-_73562046 | 4.06 |
ENSRNOT00000080501
|
Gm14597
|
predicted gene 14597 |
| chr6_-_143195445 | 4.05 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr15_-_54528480 | 4.05 |
ENSRNOT00000066888
|
Fndc3a
|
fibronectin type III domain containing 3a |
| chr4_-_21920651 | 3.98 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr17_-_42031265 | 3.95 |
ENSRNOT00000068021
|
Dcdc2
|
doublecortin domain containing 2 |
| chr14_+_23405717 | 3.94 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr1_+_169115981 | 3.93 |
ENSRNOT00000067478
|
Olr135
|
olfactory receptor 135 |
| chr5_+_153976535 | 3.91 |
ENSRNOT00000030881
|
LOC500567
|
similar to RIKEN cDNA 1700029M20 |
| chr2_+_80269661 | 3.89 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chrX_+_84064427 | 3.82 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr8_-_96547568 | 3.81 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr3_-_11417546 | 3.78 |
ENSRNOT00000018776
|
Lcn2
|
lipocalin 2 |
| chr3_-_102484517 | 3.78 |
ENSRNOT00000006522
|
Olr750
|
olfactory receptor 750 |
| chr3_-_15278645 | 3.76 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr10_+_1834518 | 3.74 |
ENSRNOT00000061709
|
Gm1758
|
predicted gene 1758 |
| chr8_-_107490093 | 3.64 |
ENSRNOT00000046832
|
LOC684466
|
similar to Fas apoptotic inhibitory molecule 1 (rFAIM) |
| chr8_+_42001912 | 3.55 |
ENSRNOT00000071709
|
Olr1226
|
olfactory receptor 1226 |
| chr1_-_252100759 | 3.55 |
ENSRNOT00000028098
|
Rnls
|
renalase, FAD-dependent amine oxidase |
| chr3_-_57607683 | 3.50 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr1_-_57518458 | 3.48 |
ENSRNOT00000002040
|
Pdcd2
|
programmed cell death 2 |
| chr17_+_85914410 | 3.46 |
ENSRNOT00000075188
|
Armc3
|
armadillo repeat containing 3 |
| chr4_-_176606382 | 3.42 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr16_+_75083172 | 3.41 |
ENSRNOT00000058002
|
Defb37
|
defensin beta 37 |
| chr17_-_89923423 | 3.41 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr2_-_157035483 | 3.39 |
ENSRNOT00000076491
|
LOC304239
|
similar to RalA binding protein 1 |
| chr6_-_104290579 | 3.38 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr13_+_91054974 | 3.38 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr9_+_66058047 | 3.37 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr4_+_2053712 | 3.35 |
ENSRNOT00000045086
|
Rnf32
|
ring finger protein 32 |
| chr1_+_255579474 | 3.33 |
ENSRNOT00000024465
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr3_-_153114520 | 3.32 |
ENSRNOT00000008254
|
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr10_+_87759769 | 3.27 |
ENSRNOT00000017378
ENSRNOT00000046526 |
Krtap9-1
|
keratin associated protein 9-1 |
| chr2_+_93669765 | 3.26 |
ENSRNOT00000045438
|
Slc10a5
|
solute carrier family 10, member 5 |
| chr8_-_8524643 | 3.25 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr3_+_21764377 | 3.25 |
ENSRNOT00000063938
|
Gpr21
|
G protein-coupled receptor 21 |
| chr3_-_45169118 | 3.24 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr2_-_60657712 | 3.19 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr6_+_18880737 | 3.19 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr3_+_81287242 | 3.16 |
ENSRNOT00000086530
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr17_-_78812111 | 3.10 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr16_+_7035068 | 3.03 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chr12_-_35979193 | 3.02 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr4_+_147832136 | 3.00 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr10_-_65502936 | 3.00 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr5_-_133959447 | 2.96 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr5_-_78324278 | 2.88 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr2_-_219262901 | 2.84 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr10_-_74413745 | 2.80 |
ENSRNOT00000038296
|
Prr11
|
proline rich 11 |
| chr2_+_96439286 | 2.77 |
ENSRNOT00000016091
|
Il7
|
interleukin 7 |
| chr2_-_149432106 | 2.72 |
ENSRNOT00000051027
|
P2ry13
|
purinergic receptor P2Y13 |
| chr8_-_83421612 | 2.69 |
ENSRNOT00000015824
|
Hcrtr2
|
hypocretin receptor 2 |
| chr8_-_43304560 | 2.67 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr2_-_198706428 | 2.63 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr4_-_22424862 | 2.59 |
ENSRNOT00000082359
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr1_+_98414226 | 2.56 |
ENSRNOT00000090785
|
Siglecl1
|
SIGLEC family like 1 |
| chr12_-_45116665 | 2.51 |
ENSRNOT00000089043
|
Taok3
|
TAO kinase 3 |
| chr18_-_69944632 | 2.42 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr6_+_104291071 | 2.41 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr8_+_113936991 | 2.37 |
ENSRNOT00000017743
|
Aste1
|
asteroid homolog 1 (Drosophila) |
| chr20_+_3155652 | 2.29 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr1_+_217345545 | 2.25 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr13_-_83457888 | 2.25 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr6_+_125876109 | 2.20 |
ENSRNOT00000091300
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr3_+_128155069 | 2.19 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr5_+_135574172 | 2.18 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr8_-_83280888 | 2.17 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chr5_+_155934490 | 2.16 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr11_+_68633160 | 2.12 |
ENSRNOT00000063795
|
Sec22a
|
SEC22 homolog A, vesicle trafficking protein |
| chr1_+_224998172 | 2.12 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chrX_+_88298266 | 2.08 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr1_+_15834779 | 2.08 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr1_-_78180216 | 2.06 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr1_-_167971151 | 2.03 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr5_+_154311998 | 2.03 |
ENSRNOT00000084085
|
Gale
|
UDP-galactose-4-epimerase |
| chr10_+_43601689 | 1.99 |
ENSRNOT00000029238
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr2_-_192027225 | 1.99 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr1_+_219390523 | 1.99 |
ENSRNOT00000054852
|
Gpr152
|
G protein-coupled receptor 152 |
| chr10_+_11046221 | 1.96 |
ENSRNOT00000005109
|
Nmral1
|
NmrA like redox sensor 1 |
| chr9_+_66888393 | 1.95 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr1_+_80293566 | 1.94 |
ENSRNOT00000024246
|
Ercc2
|
ERCC excision repair 2, TFIIH core complex helicase subunit |
| chr2_+_220432037 | 1.87 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr6_+_104291340 | 1.85 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr16_+_74785281 | 1.77 |
ENSRNOT00000064316
ENSRNOT00000017106 ENSRNOT00000090646 |
Nek3
|
NIMA-related kinase 3 |
| chr5_+_134927609 | 1.77 |
ENSRNOT00000018506
|
Lrrc41
|
leucine rich repeat containing 41 |
| chr17_-_44538780 | 1.75 |
ENSRNOT00000085743
|
AABR07027809.1
|
|
| chr2_+_195996521 | 1.73 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr5_-_12172009 | 1.72 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr10_-_87286387 | 1.71 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr3_+_147952879 | 1.69 |
ENSRNOT00000031922
|
Defb20
|
defensin beta 20 |
| chr16_+_59564419 | 1.67 |
ENSRNOT00000083434
|
Lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr11_-_43022565 | 1.67 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr15_-_88670349 | 1.67 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr13_-_90405591 | 1.63 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr3_+_19366370 | 1.61 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr14_+_76732650 | 1.58 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr10_-_34301197 | 1.56 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr5_-_162442968 | 1.50 |
ENSRNOT00000048366
|
Pramef17
|
PRAME family member 17 |
| chr6_+_132510757 | 1.50 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr1_-_197801634 | 1.46 |
ENSRNOT00000090200
|
Nfatc2ip
|
nuclear factor of activated T-cells 2 interacting protein |
| chr10_-_89005213 | 1.46 |
ENSRNOT00000027159
|
Psmc3ip
|
PSMC3 interacting protein |
| chr4_+_1658278 | 1.45 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr1_-_167770210 | 1.45 |
ENSRNOT00000024948
|
Olr47
|
olfactory receptor 47 |
| chr17_-_27279260 | 1.40 |
ENSRNOT00000018508
|
Snrnp48
|
small nuclear ribonucleoprotein U11/U12 subunit 48 |
| chr1_-_198354466 | 1.38 |
ENSRNOT00000027094
|
Tmem219
|
transmembrane protein 219 |
| chr1_-_261179790 | 1.36 |
ENSRNOT00000074420
ENSRNOT00000072073 |
Exosc1
|
exosome component 1 |
| chr10_-_66848388 | 1.36 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr16_-_32439421 | 1.32 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chr7_-_8060373 | 1.31 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr4_-_55011415 | 1.30 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr20_+_3176107 | 1.29 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr18_-_16543992 | 1.29 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr6_-_127319362 | 1.27 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr5_-_151473750 | 1.22 |
ENSRNOT00000011357
|
Tmem222
|
transmembrane protein 222 |
| chr1_+_59156251 | 1.21 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0010034 | response to acetate(GO:0010034) |
| 2.9 | 8.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 2.4 | 7.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 2.4 | 7.1 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 1.9 | 5.7 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 1.9 | 7.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.7 | 8.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.7 | 8.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.6 | 4.7 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 1.3 | 5.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.3 | 3.8 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.2 | 7.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 1.0 | 12.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.9 | 4.3 | GO:0002018 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.9 | 3.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.8 | 6.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.8 | 9.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.7 | 6.7 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.7 | 2.2 | GO:0035723 | interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) regulation of monocyte extravasation(GO:2000437) |
| 0.7 | 5.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.7 | 2.1 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.7 | 5.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.6 | 2.6 | GO:1990963 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.6 | 1.9 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.6 | 3.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.6 | 3.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.6 | 9.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.6 | 2.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.6 | 3.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 | 1.6 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.5 | 3.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.5 | 5.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.5 | 10.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.4 | 4.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 1.3 | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.4 | 4.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 7.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.4 | 3.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 3.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.4 | 1.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.4 | 3.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 3.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.4 | 2.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 2.7 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.3 | 6.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 3.3 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 7.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 1.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.3 | 3.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.3 | 2.8 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.3 | 0.8 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.3 | 7.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 0.5 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.3 | 3.2 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.3 | 2.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.3 | 2.3 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.2 | 7.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 4.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.2 | 4.5 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 2.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 2.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 4.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 0.8 | GO:0035772 | interleukin-13-mediated signaling pathway(GO:0035772) |
| 0.2 | 6.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 5.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 1.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 1.9 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.2 | 1.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 1.7 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 7.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.2 | 1.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 1.1 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 5.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 4.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 3.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 13.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.8 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 1.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 4.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 4.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 3.4 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 3.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 5.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.2 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 3.3 | GO:0046627 | positive regulation of multicellular organism growth(GO:0040018) negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 5.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 2.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 5.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 6.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.1 | 2.2 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 2.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 1.4 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.1 | 0.3 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 3.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.6 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 1.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.7 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 2.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 1.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 6.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 4.9 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 4.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 2.1 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.3 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 2.2 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 6.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 5.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 4.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 3.5 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 7.1 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.7 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.0 | 0.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 1.7 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.6 | 9.7 | GO:0071547 | piP-body(GO:0071547) |
| 1.2 | 8.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.0 | 5.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.8 | 3.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.6 | 8.8 | GO:1990023 | mitotic spindle pole(GO:0097431) mitotic spindle midzone(GO:1990023) |
| 0.5 | 1.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.5 | 4.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 1.3 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.4 | 12.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 3.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.3 | 5.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 1.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.3 | 5.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 6.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 3.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 4.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 5.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 3.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 7.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 6.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 3.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 2.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 3.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 1.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 8.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 2.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 2.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 3.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 18.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 15.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 10.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 3.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 8.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 3.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 7.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 2.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 21.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 3.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 6.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 3.0 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 15.3 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 3.7 | 14.9 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 2.9 | 8.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 2.5 | 7.4 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 2.3 | 13.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.8 | 5.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.7 | 5.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 1.7 | 5.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.4 | 7.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 1.3 | 9.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 1.1 | 3.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.0 | 3.0 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.9 | 27.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.8 | 10.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.8 | 3.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.8 | 4.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.8 | 5.3 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.7 | 8.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.7 | 2.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.7 | 6.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.7 | 6.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.7 | 12.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.6 | 3.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.6 | 1.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.6 | 8.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 3.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 2.6 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.5 | 4.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 6.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.5 | 1.9 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 3.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.4 | 1.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 3.0 | GO:0016918 | retinal binding(GO:0016918) |
| 0.4 | 7.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 4.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 7.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.3 | 1.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 2.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 1.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.2 | 4.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 4.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 5.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 1.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 2.3 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.2 | 2.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 3.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 3.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 11.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 3.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.3 | GO:0046703 | T cell receptor binding(GO:0042608) natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.0 | GO:1990446 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.1 | 1.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 8.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 2.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 3.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 5.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 11.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 2.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 3.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.4 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 3.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 4.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 3.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 17.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 11.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.8 | 13.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 17.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 8.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 3.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 6.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 2.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 2.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 5.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 8.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 6.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 3.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 5.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 6.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 5.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 12.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |