Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
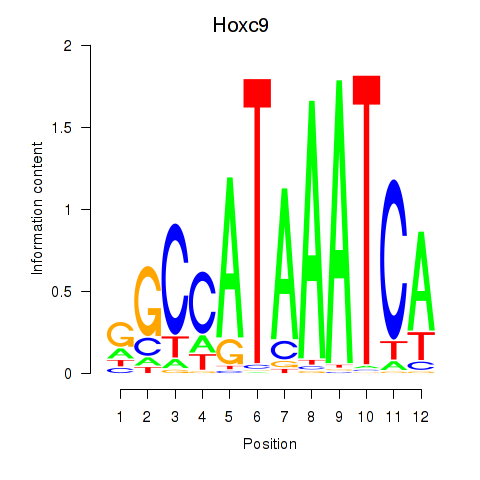
Results for Hoxc9
Z-value: 1.09
Transcription factors associated with Hoxc9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc9
|
ENSRNOG00000016199 | homeobox C9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc9 | rn6_v1_chr7_+_144595960_144595960 | 0.43 | 4.2e-16 | Click! |
Activity profile of Hoxc9 motif
Sorted Z-values of Hoxc9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_26999795 | 53.50 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr7_-_29171783 | 50.68 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr19_+_50848736 | 45.04 |
ENSRNOT00000077053
|
Cdh13
|
cadherin 13 |
| chr10_-_8498422 | 43.99 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr17_-_78735324 | 37.71 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_-_215838209 | 37.68 |
ENSRNOT00000050760
|
Igf2
|
insulin-like growth factor 2 |
| chr1_+_167870452 | 34.32 |
ENSRNOT00000025027
|
Olr63
|
olfactory receptor 63 |
| chr8_+_64481172 | 34.11 |
ENSRNOT00000015332
|
Pkm
|
pyruvate kinase, muscle |
| chr13_+_106463368 | 32.37 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chr7_-_130120579 | 31.89 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chrX_-_142248369 | 27.93 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr8_+_119228612 | 27.40 |
ENSRNOT00000078439
ENSRNOT00000043737 |
Lrrc2
|
leucine rich repeat containing 2 |
| chr3_-_23020441 | 25.73 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr12_+_27155587 | 22.06 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr20_-_25826658 | 21.08 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr1_+_201055644 | 20.98 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr16_+_8207223 | 18.74 |
ENSRNOT00000026751
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr16_+_22361998 | 18.70 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chrX_+_119390013 | 18.59 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr13_-_36290531 | 18.44 |
ENSRNOT00000071388
|
Steap3
|
STEAP3 metalloreductase |
| chrX_+_136460215 | 17.02 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr8_+_130350510 | 16.45 |
ENSRNOT00000073376
|
Ss18l2
|
SS18 like 2 |
| chr19_-_37427989 | 15.32 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_-_56683731 | 15.14 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr18_+_61261418 | 14.71 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr10_-_85974644 | 13.88 |
ENSRNOT00000006098
ENSRNOT00000082974 |
Cacnb1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr2_+_233602732 | 13.10 |
ENSRNOT00000044232
|
Pitx2
|
paired-like homeodomain 2 |
| chr2_-_96032722 | 12.95 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr7_+_123578878 | 12.20 |
ENSRNOT00000011316
|
Smdt1
|
single-pass membrane protein with aspartate-rich tail 1 |
| chr8_+_36625733 | 11.11 |
ENSRNOT00000016248
|
Cdon
|
cell adhesion associated, oncogene regulated |
| chr1_+_201620642 | 10.88 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr9_-_52238564 | 8.76 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr3_+_14889510 | 8.68 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr11_-_60679555 | 8.46 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr15_+_34234755 | 7.59 |
ENSRNOT00000059987
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr15_+_34234193 | 7.28 |
ENSRNOT00000077584
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_+_41323194 | 6.68 |
ENSRNOT00000026350
|
Esr1
|
estrogen receptor 1 |
| chr6_-_25507073 | 6.06 |
ENSRNOT00000061176
|
Plb1
|
phospholipase B1 |
| chr1_-_7480825 | 5.02 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr6_-_123577695 | 4.79 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr1_+_79754587 | 4.63 |
ENSRNOT00000083211
|
Micb
|
MHC class I polypeptide-related sequence B |
| chr3_-_103128243 | 4.26 |
ENSRNOT00000046310
|
Olr779
|
olfactory receptor 779 |
| chr3_-_103400272 | 4.05 |
ENSRNOT00000046527
|
Olr790
|
olfactory receptor 790 |
| chr5_+_154489590 | 4.02 |
ENSRNOT00000035788
|
Id3
|
inhibitor of DNA binding 3, HLH protein |
| chr10_+_103874383 | 3.96 |
ENSRNOT00000038935
|
Otop2
|
otopetrin 2 |
| chr18_-_24823837 | 3.38 |
ENSRNOT00000021405
ENSRNOT00000090390 |
Myo7b
|
myosin VIIb |
| chr1_-_140860882 | 3.25 |
ENSRNOT00000067889
ENSRNOT00000024253 ENSRNOT00000079100 |
Mfge8
|
milk fat globule-EGF factor 8 protein |
| chr3_-_103447716 | 3.09 |
ENSRNOT00000074196
|
Olr791
|
olfactory receptor 791 |
| chr5_-_128839433 | 3.04 |
ENSRNOT00000065280
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr3_-_103152122 | 3.02 |
ENSRNOT00000045743
|
Olr781
|
olfactory receptor 781 |
| chr2_+_23257915 | 2.96 |
ENSRNOT00000080942
|
AABR07007675.1
|
|
| chrX_+_15113878 | 2.94 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr15_-_36321572 | 2.89 |
ENSRNOT00000073934
|
AABR07018016.1
|
|
| chr13_-_61306939 | 2.88 |
ENSRNOT00000086610
|
Rgs21
|
regulator of G-protein signaling 21 |
| chr3_-_103485806 | 2.71 |
ENSRNOT00000072524
|
Olr789
|
olfactory receptor 789 |
| chr2_-_236149971 | 2.33 |
ENSRNOT00000089472
|
AABR07013412.1
|
|
| chr4_-_119399130 | 2.27 |
ENSRNOT00000042796
|
Vom1r101
|
vomeronasal 1 receptor 101 |
| chr8_-_63291966 | 2.15 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr6_-_33691301 | 2.00 |
ENSRNOT00000008008
|
Rhob
|
ras homolog family member B |
| chr1_-_200548317 | 2.00 |
ENSRNOT00000088610
|
AABR07005806.1
|
|
| chr1_-_155955173 | 1.82 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr7_+_42304534 | 1.58 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr20_-_27083410 | 1.58 |
ENSRNOT00000000432
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chr17_+_41798783 | 1.36 |
ENSRNOT00000023519
|
Nrsn1
|
neurensin 1 |
| chr4_+_133540540 | 0.92 |
ENSRNOT00000073232
|
AABR07061556.1
|
|
| chr2_+_66940057 | 0.64 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr11_-_37253776 | 0.53 |
ENSRNOT00000002215
|
Dscam
|
DS cell adhesion molecule |
| chr4_-_18396035 | 0.48 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr10_-_88107232 | 0.44 |
ENSRNOT00000019330
ENSRNOT00000076770 |
Krt9
|
keratin 9 |
| chr2_-_140334912 | 0.24 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr2_-_173087648 | 0.22 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr15_-_27001826 | 0.20 |
ENSRNOT00000016693
|
Olr1608
|
olfactory receptor 1608 |
| chr4_-_85329362 | 0.18 |
ENSRNOT00000014925
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.7 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 6.2 | 18.6 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) regulation of metanephros size(GO:0035566) |
| 6.1 | 18.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 5.6 | 45.0 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 5.4 | 37.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 5.1 | 50.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 4.9 | 53.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 4.4 | 13.1 | GO:0021539 | subthalamus development(GO:0021539) deltoid tuberosity development(GO:0035993) pulmonary vein morphogenesis(GO:0060577) |
| 4.0 | 27.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 3.7 | 18.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 3.4 | 34.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 2.6 | 21.1 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 2.2 | 11.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 2.2 | 8.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 2.2 | 8.7 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 1.8 | 44.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.7 | 6.7 | GO:1990375 | baculum development(GO:1990375) |
| 1.6 | 21.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 1.2 | 12.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.8 | 37.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.8 | 32.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.7 | 3.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.6 | 6.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.5 | 10.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 3.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 1.6 | GO:0070668 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 13.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 2.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 4.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 15.3 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.2 | 0.5 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 | 4.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.2 | 15.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 2.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 13.2 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.1 | 1.6 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 0.5 | GO:0021886 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) cranial ganglion development(GO:0061550) trigeminal ganglion development(GO:0061551) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 2.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.1 | 8.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 51.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 4.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 13.0 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 18.7 | GO:0055114 | oxidation-reduction process(GO:0055114) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 50.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 2.9 | 8.7 | GO:1990032 | parallel fiber(GO:1990032) |
| 2.2 | 8.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.0 | 12.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.1 | 21.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.7 | 6.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.6 | 15.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.6 | 10.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 13.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.5 | 14.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.5 | 18.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.5 | 8.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 27.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.4 | 44.0 | GO:0005901 | caveola(GO:0005901) |
| 0.4 | 13.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 53.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 4.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 25.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 30.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 34.1 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.1 | 6.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 18.7 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 13.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 13.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 43.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 47.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 75.5 | GO:0005886 | plasma membrane(GO:0005886) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 45.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 8.5 | 34.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 8.1 | 32.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 6.5 | 104.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 6.2 | 18.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 4.6 | 18.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 3.7 | 18.6 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 2.3 | 21.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 2.0 | 6.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 1.6 | 10.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.4 | 8.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.4 | 31.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.3 | 6.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.1 | 31.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.7 | 13.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 27.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.5 | 13.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 25.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 3.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 13.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 8.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 1.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 31.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.2 | 4.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 5.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 21.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.2 | 41.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.2 | 8.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 16.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 4.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 5.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 51.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 10.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 18.7 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 2.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 31.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 36.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.5 | 11.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 13.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 15.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 8.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 8.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 18.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 21.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 10.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 31.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 1.8 | 37.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.3 | 50.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.3 | 36.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.9 | 64.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.8 | 18.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 18.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.4 | 11.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 16.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 15.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 21.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 11.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |