Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxc6
Z-value: 0.36
Transcription factors associated with Hoxc6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc6
|
ENSRNOG00000016442 | homeo box C6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc6 | rn6_v1_chr7_+_144623555_144623555 | 0.46 | 6.8e-18 | Click! |
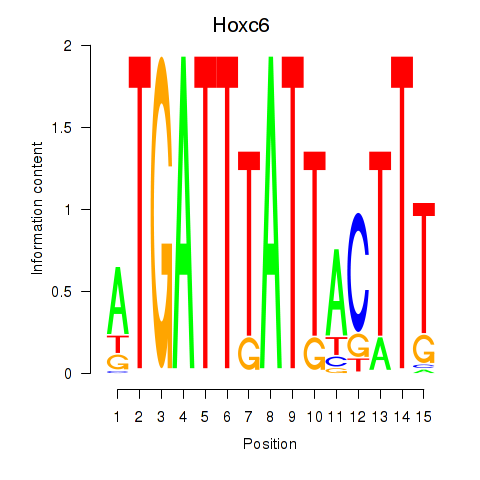
Activity profile of Hoxc6 motif
Sorted Z-values of Hoxc6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_78793336 | 12.71 |
ENSRNOT00000057898
|
Mt1
|
metallothionein 1 |
| chr10_-_59883839 | 12.25 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr5_-_79008363 | 10.61 |
ENSRNOT00000010040
|
Kif12
|
kinesin family member 12 |
| chr20_-_14025067 | 9.08 |
ENSRNOT00000093430
ENSRNOT00000074533 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr9_+_25410669 | 7.79 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr9_-_4685383 | 7.70 |
ENSRNOT00000061896
|
Sult1c2
|
sulfotransferase family 1C member 2 |
| chr4_-_176381477 | 6.98 |
ENSRNOT00000048367
|
Slco1a6
|
solute carrier organic anion transporter family, member 1a6 |
| chr1_+_229063714 | 6.85 |
ENSRNOT00000087526
|
Glyat
|
glycine-N-acyltransferase |
| chr19_-_62150423 | 5.42 |
ENSRNOT00000073524
ENSRNOT00000091209 |
Ccdc7
|
coiled-coil domain containing 7 |
| chr13_-_80968971 | 4.72 |
ENSRNOT00000004931
|
Mroh9
|
maestro heat-like repeat family member 9 |
| chr5_+_147257678 | 4.65 |
ENSRNOT00000066268
|
Rnf19b
|
ring finger protein 19B |
| chr6_-_86223052 | 4.48 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr2_-_205426869 | 4.40 |
ENSRNOT00000023333
|
Sycp1
|
synaptonemal complex protein 1 |
| chr11_-_68349238 | 4.13 |
ENSRNOT00000030074
|
Sema5b
|
semaphorin 5B |
| chr4_-_11497531 | 3.60 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_-_145062956 | 3.37 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr8_+_115193146 | 3.21 |
ENSRNOT00000056391
|
Iqcf1
|
IQ motif containing F1 |
| chr6_+_48452369 | 2.49 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr13_-_73055631 | 2.46 |
ENSRNOT00000081892
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chrX_-_115175299 | 2.31 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr14_+_91557601 | 2.25 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr2_-_120316600 | 2.15 |
ENSRNOT00000084521
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr1_+_101541266 | 2.09 |
ENSRNOT00000028436
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr4_-_165118062 | 1.96 |
ENSRNOT00000087219
|
LOC502908
|
Ly49s8 |
| chr7_-_55604403 | 1.96 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr4_+_79021872 | 1.85 |
ENSRNOT00000012677
ENSRNOT00000067125 |
Fam221a
|
family with sequence similarity 221, member A |
| chr3_-_102432479 | 1.82 |
ENSRNOT00000047270
|
Olr747
|
olfactory receptor 747 |
| chr20_-_31402483 | 1.82 |
ENSRNOT00000033529
|
H2afy2
|
H2A histone family, member Y2 |
| chr14_-_82171480 | 1.81 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chrX_+_141847092 | 1.69 |
ENSRNOT00000004684
|
Magea11
|
melanoma antigen family A, 11 |
| chr4_+_101909389 | 1.64 |
ENSRNOT00000086458
|
AABR07060963.2
|
|
| chr3_+_173955686 | 1.54 |
ENSRNOT00000088447
|
Fam217b
|
family with sequence similarity 217, member B |
| chr1_+_128606770 | 1.50 |
ENSRNOT00000073355
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr7_+_11660934 | 1.47 |
ENSRNOT00000022336
|
Lmnb2
|
lamin B2 |
| chr4_+_98481520 | 1.45 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr2_-_105089659 | 1.32 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr1_-_276012351 | 1.31 |
ENSRNOT00000045642
|
AABR07007000.1
|
|
| chr4_-_82173207 | 1.30 |
ENSRNOT00000074167
|
Hoxa5
|
homeo box A5 |
| chr3_-_113342675 | 1.28 |
ENSRNOT00000020130
|
Strc
|
stereocilin |
| chr2_-_140334912 | 1.21 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr7_+_34402738 | 1.18 |
ENSRNOT00000030985
|
Ccdc38
|
coiled-coil domain containing 38 |
| chr12_-_44911147 | 1.16 |
ENSRNOT00000071074
ENSRNOT00000046190 |
Ksr2
|
kinase suppressor of ras 2 |
| chr13_+_89386023 | 1.14 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr11_+_45888221 | 1.13 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr14_+_9555264 | 1.11 |
ENSRNOT00000002928
|
Nkx6-1
|
NK6 homeobox 1 |
| chr13_+_89385859 | 1.11 |
ENSRNOT00000047434
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr5_+_10178302 | 1.10 |
ENSRNOT00000009679
|
Sntg1
|
syntrophin, gamma 1 |
| chrX_+_96863891 | 1.10 |
ENSRNOT00000085665
|
AABR07040284.1
|
|
| chr19_-_38484611 | 1.04 |
ENSRNOT00000077960
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr12_+_27155587 | 0.92 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr10_+_46018659 | 0.91 |
ENSRNOT00000040865
ENSRNOT00000004357 |
Mprip
|
myosin phosphatase Rho interacting protein |
| chr1_-_172665244 | 0.87 |
ENSRNOT00000089570
|
Olr264
|
olfactory receptor 264 |
| chr4_-_58006839 | 0.77 |
ENSRNOT00000076645
|
Cep41
|
centrosomal protein 41 |
| chr8_+_116307141 | 0.69 |
ENSRNOT00000037375
|
Rassf1
|
Ras association domain family member 1 |
| chr2_-_23909016 | 0.58 |
ENSRNOT00000084044
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr1_+_227757425 | 0.57 |
ENSRNOT00000032937
ENSRNOT00000049574 |
Ms4a6bl
|
membrane-spanning 4-domains, subfamily A, member 6B-like |
| chr16_+_90325304 | 0.54 |
ENSRNOT00000057310
|
Slc10a2
|
solute carrier family 10 member 2 |
| chr17_-_77527894 | 0.51 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr8_-_87245951 | 0.50 |
ENSRNOT00000015299
|
Tmem30a
|
transmembrane protein 30A |
| chr3_+_16571602 | 0.39 |
ENSRNOT00000048351
|
LOC100361705
|
rCG64259-like |
| chr4_+_93959152 | 0.39 |
ENSRNOT00000058437
|
AABR07060795.1
|
|
| chr7_-_90318221 | 0.33 |
ENSRNOT00000050774
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr1_+_224933529 | 0.18 |
ENSRNOT00000088282
|
Wdr74
|
WD repeat domain 74 |
| chr7_-_120919261 | 0.12 |
ENSRNOT00000019612
|
Josd1
|
Josephin domain containing 1 |
| chrX_+_136460215 | 0.05 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr3_-_103485806 | 0.02 |
ENSRNOT00000072524
|
Olr789
|
olfactory receptor 789 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.1 | GO:0031179 | peptide modification(GO:0031179) |
| 2.1 | 12.7 | GO:0046687 | response to chromate(GO:0046687) |
| 1.3 | 7.8 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.8 | 3.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.6 | 4.4 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.6 | 1.8 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.6 | 12.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.5 | 1.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.5 | 3.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 1.3 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.3 | 6.9 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.3 | 2.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 | 1.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 2.5 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 7.5 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.2 | 4.7 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 4.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 2.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 2.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.9 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.1 | 1.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.1 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 1.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 10.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0000802 | transverse filament(GO:0000802) |
| 0.6 | 4.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.4 | 3.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 4.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.8 | GO:0001740 | Barr body(GO:0001740) |
| 0.2 | 1.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 10.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.3 | GO:0032426 | stereocilium tip(GO:0032426) kinocilium(GO:0060091) |
| 0.0 | 19.1 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 3.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.5 | 9.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 1.1 | 6.9 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.9 | 3.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 2.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.6 | 2.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 2.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 7.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 2.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 12.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 1.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 10.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 4.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 7.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.3 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 9.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 6.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 3.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |