Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
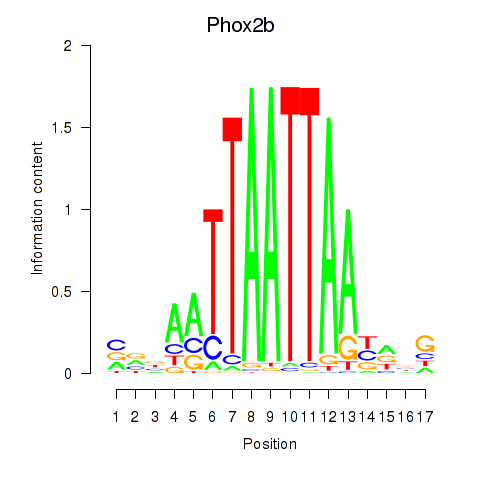
Results for Hoxc4_Arx_Otp_Esx1_Phox2b
Z-value: 0.67
Transcription factors associated with Hoxc4_Arx_Otp_Esx1_Phox2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
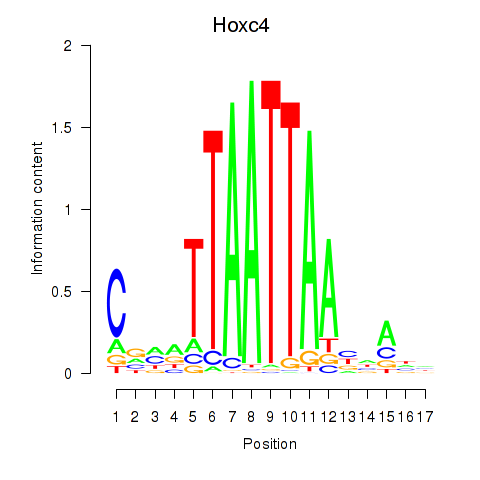
Hoxc4
|
ENSRNOG00000016613 | homeo box C4 |
|
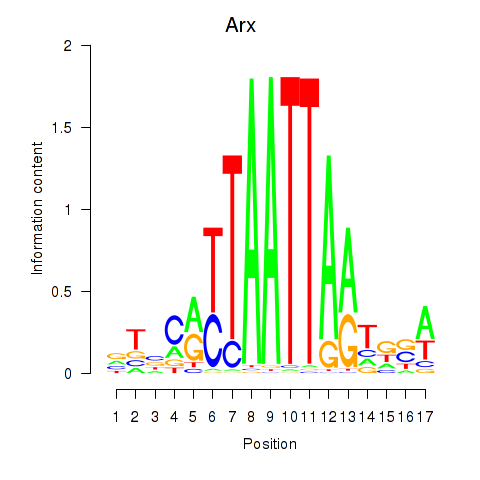
Arx
|
ENSRNOG00000053562 | aristaless related homeobox |
|
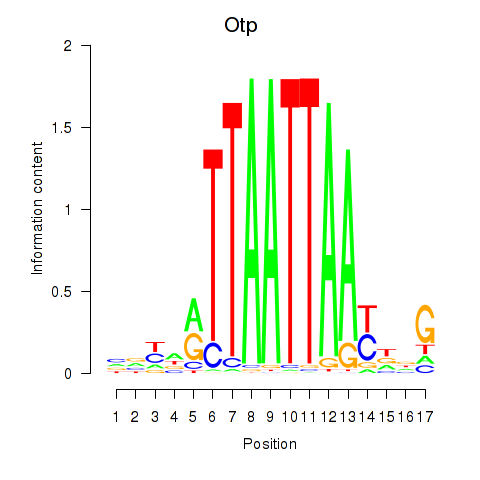
Otp
|
ENSRNOG00000010560 | orthopedia homeobox |
|
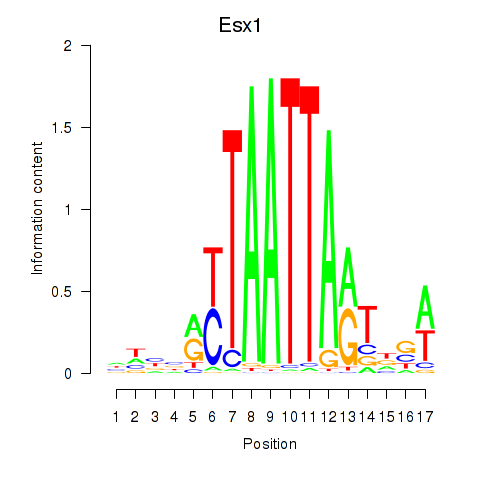
Esx1
|
ENSRNOG00000048011 | ESX homeobox 1 |
|
Phox2b
|
ENSRNOG00000051929 | paired-like homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc4 | rn6_v1_chr7_+_144647587_144647587 | 0.32 | 3.7e-09 | Click! |
| Otp | rn6_v1_chr2_+_24546536_24546536 | 0.15 | 7.8e-03 | Click! |
| Arx | rn6_v1_chrX_+_62366453_62366453 | 0.14 | 1.2e-02 | Click! |
| Esx1 | rn6_v1_chrX_-_107762590_107762590 | -0.11 | 5.8e-02 | Click! |
| Phox2b | rn6_v1_chr14_+_42714315_42714315 | 0.02 | 7.5e-01 | Click! |
Activity profile of Hoxc4_Arx_Otp_Esx1_Phox2b motif
Sorted Z-values of Hoxc4_Arx_Otp_Esx1_Phox2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_161981441 | 49.93 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr2_-_158156444 | 44.65 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_-_40086870 | 27.86 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr4_+_29535852 | 26.65 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr1_+_61786900 | 22.27 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr3_-_37854561 | 22.00 |
ENSRNOT00000076095
|
Neb
|
nebulin |
| chr3_+_159368273 | 21.74 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr17_-_87826421 | 20.90 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr2_+_158097843 | 20.88 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr2_-_158156150 | 19.14 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr4_-_18396035 | 17.84 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr17_-_79085076 | 17.03 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr4_-_72143748 | 16.57 |
ENSRNOT00000024428
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr18_-_26656879 | 16.33 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_+_37329779 | 16.11 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr20_+_34258791 | 14.36 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr7_-_76488216 | 14.05 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chrX_+_14019961 | 13.42 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr6_+_8284878 | 13.32 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr3_-_14229067 | 12.83 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr19_+_40855433 | 12.79 |
ENSRNOT00000023823
|
Il34
|
interleukin 34 |
| chr18_+_30036887 | 12.75 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr2_+_145174876 | 12.59 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chrX_+_84064427 | 11.77 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr14_-_115052450 | 11.73 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr9_-_44237117 | 11.61 |
ENSRNOT00000068496
|
RGD1310819
|
similar to putative protein (5S487) |
| chr2_-_35870578 | 11.26 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr18_-_55891710 | 11.01 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr15_+_33606124 | 10.55 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr9_+_73378057 | 10.43 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr5_-_119564846 | 10.22 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr5_+_165724027 | 10.13 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr9_-_30844199 | 9.98 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr8_-_39460844 | 9.98 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr11_-_11585078 | 9.80 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr17_+_23661429 | 9.80 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr18_+_32336102 | 9.73 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr13_-_92988137 | 9.51 |
ENSRNOT00000004803
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr11_+_36851038 | 9.35 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr14_+_34446616 | 8.42 |
ENSRNOT00000002976
|
Clock
|
clock circadian regulator |
| chr9_-_20154077 | 7.92 |
ENSRNOT00000082904
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr1_-_276228574 | 7.35 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr6_+_104291071 | 7.11 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr4_+_169161585 | 7.04 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr4_+_180291389 | 7.04 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr6_+_2216623 | 6.75 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_+_58724855 | 6.71 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr18_+_35384743 | 6.63 |
ENSRNOT00000076143
ENSRNOT00000074593 |
LOC100911797
|
serine protease inhibitor Kazal-type 5-like |
| chr1_-_224698514 | 6.60 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr5_-_7941822 | 6.59 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr18_+_30017918 | 6.50 |
ENSRNOT00000079794
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_-_45169118 | 6.47 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr14_-_45859908 | 6.21 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr5_+_145311375 | 6.20 |
ENSRNOT00000019224
|
Smim12
|
small integral membrane protein 12 |
| chr17_-_14627937 | 6.17 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr8_-_6305033 | 6.14 |
ENSRNOT00000029887
|
Cep126
|
centrosomal protein 126 |
| chr13_+_57243877 | 6.13 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr18_+_17043903 | 6.09 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr4_+_169147243 | 6.06 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr1_+_79631668 | 6.05 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr5_+_36566783 | 6.04 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr18_+_29987206 | 6.03 |
ENSRNOT00000027383
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_-_215284548 | 6.00 |
ENSRNOT00000090375
|
Cttn
|
cortactin |
| chr19_-_56983807 | 5.89 |
ENSRNOT00000043529
|
Cox6c-ps1
|
cytochrome c oxidase subunit VIc, pseudogene |
| chr2_+_202200797 | 5.75 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr18_-_60002529 | 5.75 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
| chr20_+_19325121 | 5.58 |
ENSRNOT00000058151
|
Phyhipl
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr7_+_37101391 | 5.44 |
ENSRNOT00000029764
|
Eea1
|
early endosome antigen 1 |
| chr2_+_252090669 | 5.43 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr9_+_79630604 | 5.42 |
ENSRNOT00000021539
ENSRNOT00000083979 |
Tmem169
|
transmembrane protein 169 |
| chr7_-_120077612 | 5.41 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr1_-_278042312 | 5.38 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr5_-_2982603 | 5.38 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chrX_+_15113878 | 5.38 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr5_+_58995249 | 5.36 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr6_-_142418779 | 5.25 |
ENSRNOT00000072280
ENSRNOT00000065808 |
AABR07065814.1
|
|
| chr4_-_16654811 | 5.20 |
ENSRNOT00000008637
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr5_+_28485619 | 5.15 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr5_-_17061837 | 5.13 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr14_-_77810147 | 5.02 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr20_-_13994794 | 5.02 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr3_+_77414999 | 4.97 |
ENSRNOT00000047910
|
Olr659
|
olfactory receptor 659 |
| chr16_+_84465656 | 4.95 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr1_+_7252349 | 4.87 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chrX_-_121731543 | 4.86 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr12_-_2174131 | 4.55 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_-_120076788 | 4.53 |
ENSRNOT00000047158
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr8_+_79638696 | 4.47 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr18_+_30023828 | 4.43 |
ENSRNOT00000079008
|
Pcdha4
|
protocadherin alpha 4 |
| chr13_-_102857551 | 4.43 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr8_-_84506328 | 4.32 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr1_-_236729412 | 4.28 |
ENSRNOT00000054794
|
Prune2
|
prune homolog 2 |
| chr1_-_190370499 | 4.27 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr6_+_104291340 | 4.21 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr12_-_37538403 | 4.05 |
ENSRNOT00000001402
|
Snrnp35
|
small nuclear ribonucleoprotein U11/U12 subunit 35 |
| chr2_+_248398917 | 4.02 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr9_+_51302151 | 4.01 |
ENSRNOT00000085908
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr8_-_7426611 | 4.01 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr11_-_62067655 | 3.98 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_+_63379031 | 3.90 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr5_+_114516889 | 3.85 |
ENSRNOT00000011767
|
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr17_-_84247038 | 3.83 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr14_-_16903242 | 3.75 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chrX_-_77559348 | 3.74 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr2_-_35104963 | 3.73 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr9_-_105693357 | 3.71 |
ENSRNOT00000066968
|
Nudt12
|
nudix hydrolase 12 |
| chr2_-_33025271 | 3.68 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr18_+_3887419 | 3.64 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr19_+_53044379 | 3.62 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr11_+_58624198 | 3.59 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr6_+_64789940 | 3.55 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr1_+_259958310 | 3.53 |
ENSRNOT00000019751
|
LOC103689954
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr4_+_87608301 | 3.52 |
ENSRNOT00000058702
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr5_-_147412705 | 3.46 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr7_+_63747577 | 3.42 |
ENSRNOT00000086123
ENSRNOT00000067395 ENSRNOT00000080052 |
RGD1561648
|
RGD1561648 |
| chr20_-_5020150 | 3.39 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr2_+_66940057 | 3.36 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr3_+_100769839 | 3.35 |
ENSRNOT00000077703
|
Bdnf
|
brain-derived neurotrophic factor |
| chr7_+_6644643 | 3.31 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr2_+_30685840 | 3.20 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr7_+_42304534 | 3.17 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr3_-_51643140 | 3.13 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr13_-_91735361 | 3.13 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr18_+_30840868 | 3.13 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr12_-_35979193 | 3.10 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_+_113984646 | 3.10 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr12_-_1195591 | 2.91 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr6_+_44225233 | 2.88 |
ENSRNOT00000066593
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr1_-_215033460 | 2.88 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr5_-_166116516 | 2.81 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr13_+_84474319 | 2.78 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr6_+_101532518 | 2.78 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr4_+_88694583 | 2.77 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr4_+_155321553 | 2.76 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr12_-_40590361 | 2.76 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr18_+_29951094 | 2.74 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr5_-_17061361 | 2.72 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr7_-_29171783 | 2.70 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chrX_+_73390903 | 2.68 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr16_-_3765917 | 2.67 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr1_+_252284464 | 2.62 |
ENSRNOT00000027969
|
Lipf
|
lipase F, gastric type |
| chr8_+_44136496 | 2.60 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr4_-_176909075 | 2.58 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr16_+_46731403 | 2.58 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_22281788 | 2.57 |
ENSRNOT00000021318
|
Stx7
|
syntaxin 7 |
| chr13_-_50514151 | 2.55 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr17_+_32082937 | 2.49 |
ENSRNOT00000074775
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr2_+_205160405 | 2.47 |
ENSRNOT00000035605
|
Tspan2
|
tetraspanin 2 |
| chr1_-_217631862 | 2.44 |
ENSRNOT00000078514
ENSRNOT00000079805 |
Cttn
|
cortactin |
| chr4_+_88328061 | 2.42 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr8_+_117297670 | 2.40 |
ENSRNOT00000082628
|
Qars
|
glutaminyl-tRNA synthetase |
| chr16_+_48513432 | 2.40 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr12_+_25435567 | 2.39 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr2_-_147392062 | 2.38 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr17_+_32973695 | 2.32 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr10_-_51778939 | 2.30 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr17_-_2278613 | 2.29 |
ENSRNOT00000046525
|
Cntnap3b
|
contactin associated protein-like 3B |
| chr10_+_38919167 | 2.26 |
ENSRNOT00000077569
|
Kif3a
|
kinesin family member 3a |
| chr4_-_115208026 | 2.25 |
ENSRNOT00000084388
ENSRNOT00000066678 |
Dguok
|
deoxyguanosine kinase |
| chr2_-_88660449 | 2.20 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr16_-_8885797 | 2.20 |
ENSRNOT00000073370
|
RGD1564899
|
similar to chromosome 10 open reading frame 71 |
| chr6_-_3355339 | 2.18 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr13_-_111581018 | 2.13 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr1_-_67094567 | 2.12 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr20_+_42966140 | 2.12 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chrX_-_159244879 | 2.11 |
ENSRNOT00000030445
|
Map7d3
|
MAP7 domain containing 3 |
| chr5_+_107369730 | 2.10 |
ENSRNOT00000046837
|
LOC100912314
|
interferon alpha-1-like |
| chr13_-_44540516 | 2.09 |
ENSRNOT00000005244
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr1_+_101687855 | 2.09 |
ENSRNOT00000028546
ENSRNOT00000091597 |
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr16_-_83084975 | 2.08 |
ENSRNOT00000017947
|
Arhgef7
|
Rho guanine nucleotide exchange factor 7 |
| chr3_-_154627257 | 2.07 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr3_+_14889510 | 2.07 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr5_+_153269526 | 2.07 |
ENSRNOT00000091494
|
Syf2
|
SYF2 pre-mRNA-splicing factor |
| chr4_-_62860446 | 2.05 |
ENSRNOT00000015752
|
Fam180a
|
family with sequence similarity 180, member A |
| chr2_-_5577369 | 2.04 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr17_+_53962444 | 2.02 |
ENSRNOT00000080101
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr2_+_85305225 | 1.97 |
ENSRNOT00000015904
|
Tas2r119
|
taste receptor, type 2, member 119 |
| chr16_+_6048004 | 1.97 |
ENSRNOT00000020750
|
Actr8
|
ARP8 actin-related protein 8 homolog |
| chr4_-_51199570 | 1.93 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chrX_+_111735820 | 1.89 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr9_+_73418607 | 1.83 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr4_-_41212072 | 1.82 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr16_+_10417185 | 1.80 |
ENSRNOT00000082186
|
Anxa8
|
annexin A8 |
| chr1_+_59156251 | 1.78 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr3_+_61613774 | 1.78 |
ENSRNOT00000002148
|
Hoxd10
|
homeo box D10 |
| chr3_-_57607683 | 1.78 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr20_+_44680449 | 1.77 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr10_-_89130339 | 1.77 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chrX_-_111191932 | 1.77 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr8_-_78096302 | 1.76 |
ENSRNOT00000086185
ENSRNOT00000077999 |
Myzap
|
myocardial zonula adherens protein |
| chr12_-_18120476 | 1.74 |
ENSRNOT00000080364
|
RGD1566386
|
similar to Hypothetical protein A430033K04 |
| chr9_+_64745051 | 1.74 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr4_+_61814974 | 1.73 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr15_+_87722221 | 1.71 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr14_-_84334066 | 1.70 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chrX_-_138118696 | 1.68 |
ENSRNOT00000084130
|
Frmd7
|
FERM domain containing 7 |
| chr17_+_22984844 | 1.68 |
ENSRNOT00000078310
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr16_+_7035068 | 1.67 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chr3_+_119561290 | 1.66 |
ENSRNOT00000015843
|
Blvra
|
biliverdin reductase A |
| chr2_-_96509424 | 1.65 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc4_Arx_Otp_Esx1_Phox2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 46.9 | GO:0015884 | folic acid transport(GO:0015884) |
| 5.2 | 20.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 5.2 | 20.9 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 5.2 | 15.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 4.5 | 17.8 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 3.4 | 10.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 3.2 | 9.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 2.8 | 11.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.6 | 7.9 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 2.4 | 16.6 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 2.3 | 9.4 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 2.1 | 12.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 2.0 | 10.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 2.0 | 9.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.8 | 5.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.5 | 6.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 1.4 | 5.8 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.4 | 1.4 | GO:0048252 | lauric acid metabolic process(GO:0048252) |
| 1.3 | 4.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.3 | 12.8 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 1.3 | 3.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.2 | 3.7 | GO:0019677 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 1.2 | 8.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 1.1 | 8.4 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 1.0 | 3.1 | GO:0042223 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.0 | 18.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.0 | 4.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.0 | 2.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.9 | 3.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.9 | 5.5 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.9 | 3.6 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.9 | 7.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.8 | 2.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) phenotypic switching(GO:0036166) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.8 | 2.3 | GO:2000769 | epidermal stem cell homeostasis(GO:0036334) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.7 | 3.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.7 | 2.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.7 | 2.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.7 | 10.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.7 | 1.3 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.6 | 3.2 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.6 | 5.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.6 | 11.9 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.6 | 6.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 1.7 | GO:0016488 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.6 | 2.8 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.6 | 2.8 | GO:0099628 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.5 | 2.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.5 | 2.6 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.5 | 1.5 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.5 | 1.5 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.5 | 2.5 | GO:0002003 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.5 | 0.5 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.4 | 3.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.4 | 3.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.4 | 1.7 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.4 | 0.4 | GO:1905237 | response to cyclosporin A(GO:1905237) |
| 0.4 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.4 | 3.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.4 | 6.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 12.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.4 | 1.6 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.4 | 3.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 1.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.4 | 2.7 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 2.3 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.4 | 2.6 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.4 | 1.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 3.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 5.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.4 | 4.2 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 1.0 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 1.7 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.3 | 13.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 1.8 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.3 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 2.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 1.1 | GO:0070086 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 2.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 0.8 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.3 | 0.8 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.3 | 2.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.3 | 0.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 0.5 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.3 | 5.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 36.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 5.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.3 | 14.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.7 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.2 | 0.9 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 5.7 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 5.4 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 3.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 4.3 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 29.1 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.2 | 4.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 0.7 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 | 21.7 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.2 | 14.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.2 | 2.9 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 1.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.5 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.2 | 1.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 2.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 7.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 25.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.2 | 1.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 2.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.9 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 1.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 2.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.7 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 4.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 2.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 2.1 | GO:1902358 | sulfate transport(GO:0008272) sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.4 | GO:1903595 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 1.8 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.0 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 3.4 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 5.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 2.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 2.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 1.0 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 14.6 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 2.5 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.3 | GO:0034031 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 5.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.9 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.6 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.3 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 1.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 5.4 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 2.4 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 3.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 1.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.4 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.6 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.1 | 1.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 3.4 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 0.7 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.7 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 6.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 9.8 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.1 | 2.1 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 1.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 6.1 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 7.6 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0043474 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.0 | 0.5 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 3.5 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 5.4 | GO:0016055 | Wnt signaling pathway(GO:0016055) |
| 0.0 | 1.2 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 1.0 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 2.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.9 | GO:0097480 | synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 27.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 5.4 | 16.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 3.7 | 11.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 2.8 | 49.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.7 | 5.2 | GO:0044317 | rod spherule(GO:0044317) |
| 1.6 | 11.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.4 | 8.4 | GO:0098871 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 1.4 | 8.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 1.4 | 12.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.3 | 7.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.2 | 5.8 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.0 | 4.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.9 | 5.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.7 | 2.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.7 | 2.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.6 | 2.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 3.6 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.6 | 9.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 4.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.5 | 2.1 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.5 | 4.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.5 | 3.7 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.5 | 2.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.4 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.4 | 7.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 11.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 2.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 2.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 2.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.3 | 7.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 5.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 2.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 6.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 0.7 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 2.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 11.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 3.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 3.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 27.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 6.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 3.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 16.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 2.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 0.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 3.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 13.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 46.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 7.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 11.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 20.7 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 16.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 5.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.4 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.2 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.1 | 0.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 3.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 37.0 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 7.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 7.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 2.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 21.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.5 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 12.3 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 46.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 4.3 | 12.8 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 4.3 | 12.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 4.0 | 16.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 3.4 | 10.2 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 2.8 | 19.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.8 | 3.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.6 | 6.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.5 | 19.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.4 | 25.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.3 | 7.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.2 | 6.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.0 | 4.0 | GO:0019002 | GMP binding(GO:0019002) |
| 1.0 | 9.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.9 | 3.7 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.9 | 9.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.9 | 2.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.8 | 5.0 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.8 | 13.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 2.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.7 | 5.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.7 | 6.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.6 | 13.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 1.7 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.6 | 2.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.5 | 9.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 3.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.5 | 14.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.5 | 1.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.5 | 2.8 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.5 | 3.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 5.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 4.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 3.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.4 | 2.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 10.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.4 | 25.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 1.5 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 11.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 2.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 2.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 4.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 2.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 2.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 6.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 7.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 11.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 5.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 0.8 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 5.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 2.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 16.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 6.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 5.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 6.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.2 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 6.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 17.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 8.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.2 | 2.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 3.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 22.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 3.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 7.9 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 1.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 4.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 34.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.4 | GO:0016671 | peptide disulfide oxidoreductase activity(GO:0015037) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.3 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.1 | 25.1 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 8.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 3.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 7.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 29.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 4.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0022843 | voltage-gated cation channel activity(GO:0022843) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 2.2 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 2.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 2.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 3.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 5.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 3.8 | GO:0046873 | metal ion transmembrane transporter activity(GO:0046873) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 19.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.5 | 8.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 7.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 3.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 12.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 8.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 9.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 22.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 6.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 8.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 6.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 3.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 4.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 5.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 10.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 19.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.1 | 16.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.9 | 14.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.7 | 9.7 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.6 | 24.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 6.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 12.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 8.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 6.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 9.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 2.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 9.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 4.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 6.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 5.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 6.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 2.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 3.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 5.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 6.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 5.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 1.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 4.8 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |