Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
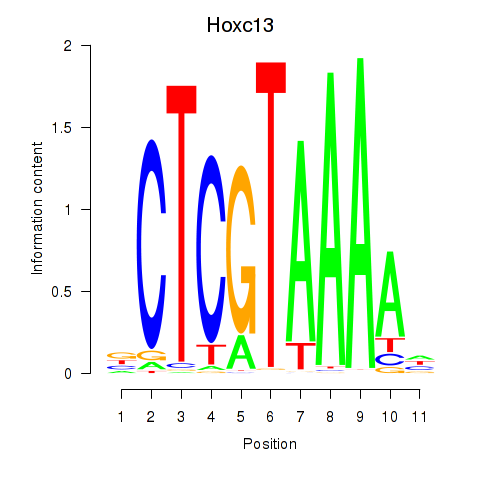
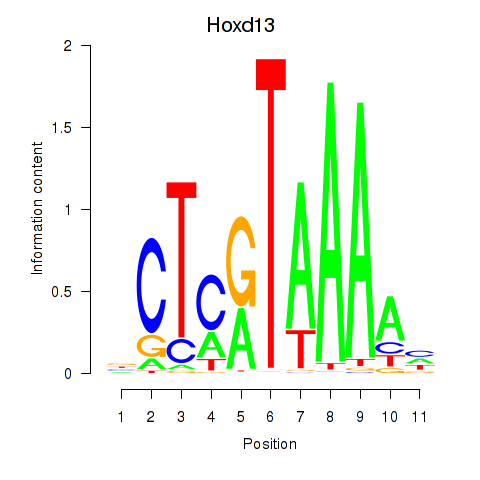
Results for Hoxc13_Hoxd13
Z-value: 0.51
Transcription factors associated with Hoxc13_Hoxd13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc13
|
ENSRNOG00000028679 | homeobox C13 |
|
Hoxd13
|
ENSRNOG00000001588 | homeo box D13 |
Activity profile of Hoxc13_Hoxd13 motif
Sorted Z-values of Hoxc13_Hoxd13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_61070599 | 14.56 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr1_-_2627747 | 12.94 |
ENSRNOT00000022003
|
Ust
|
uronyl-2-sulfotransferase |
| chr9_+_67699379 | 12.91 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr4_+_98337367 | 12.36 |
ENSRNOT00000042165
|
AABR07060872.1
|
|
| chr4_-_159192526 | 11.98 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr7_-_143228060 | 11.61 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr12_-_21760292 | 9.33 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chrX_-_77675487 | 9.21 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr16_-_61091169 | 8.72 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr6_+_96834525 | 7.29 |
ENSRNOT00000077935
|
Hif1a
|
hypoxia inducible factor 1 alpha subunit |
| chr10_-_88533829 | 6.97 |
ENSRNOT00000080521
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr1_-_271275989 | 6.24 |
ENSRNOT00000075570
|
LOC100912218
|
inositol 1,4,5-trisphosphate receptor-interacting protein-like |
| chr6_-_41039437 | 6.17 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr5_-_154332940 | 5.69 |
ENSRNOT00000014458
|
Pithd1
|
PITH domain containing 1 |
| chr1_-_43638161 | 5.67 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_143453544 | 5.52 |
ENSRNOT00000034450
ENSRNOT00000083956 |
Krt1
Krt5
|
keratin 1 keratin 5 |
| chr9_+_10044756 | 5.50 |
ENSRNOT00000071188
|
Pspn
|
persephin |
| chr10_-_29196563 | 5.10 |
ENSRNOT00000005205
|
Fabp6
|
fatty acid binding protein 6 |
| chr1_-_206282575 | 5.09 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr3_-_165700489 | 5.01 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr20_-_27578244 | 4.80 |
ENSRNOT00000000708
|
Fam26d
|
family with sequence similarity 26, member D |
| chr8_+_96551245 | 3.94 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr10_-_87954055 | 3.90 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr3_-_154490851 | 3.67 |
ENSRNOT00000017213
|
Tti1
|
TELO2 interacting protein 1 |
| chr10_+_108132105 | 3.65 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr5_-_146795866 | 3.59 |
ENSRNOT00000065640
|
Tlr12
|
toll-like receptor 12 |
| chr20_+_37876650 | 3.43 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr10_-_88009576 | 3.18 |
ENSRNOT00000018566
|
Krt35
|
keratin 35 |
| chr5_+_122019301 | 3.13 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr17_-_84247038 | 3.09 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr8_-_46694613 | 3.06 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr10_+_42614713 | 3.02 |
ENSRNOT00000081136
ENSRNOT00000073148 |
Gria1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chr3_+_110574417 | 3.01 |
ENSRNOT00000031231
|
Disp2
|
dispatched RND transporter family member 2 |
| chr15_-_34392066 | 3.00 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr1_-_84353725 | 2.99 |
ENSRNOT00000057177
|
Pld3
|
phospholipase D family, member 3 |
| chr15_+_41643541 | 2.98 |
ENSRNOT00000019646
|
Arl11
|
ADP-ribosylation factor like GTPase 11 |
| chr17_-_11037738 | 2.90 |
ENSRNOT00000043796
|
AABR07027015.1
|
|
| chr8_+_122197027 | 2.85 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr19_-_19315357 | 2.70 |
ENSRNOT00000018888
|
Cyld
|
CYLD lysine 63 deubiquitinase |
| chr4_+_140247313 | 2.47 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr7_-_143085833 | 2.43 |
ENSRNOT00000031257
ENSRNOT00000077286 |
Krt83
|
keratin 83 |
| chr6_+_123034304 | 2.28 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr7_-_140437467 | 2.20 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr15_-_37831031 | 2.20 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr2_+_40554146 | 1.94 |
ENSRNOT00000015138
|
Pde4d
|
phosphodiesterase 4D |
| chr5_-_125345726 | 1.90 |
ENSRNOT00000011244
|
LOC688684
|
similar to 60S ribosomal protein L32 |
| chr4_-_82186190 | 1.86 |
ENSRNOT00000071729
|
LOC100911622
|
homeobox protein Hox-A7-like |
| chr6_-_104289668 | 1.83 |
ENSRNOT00000085646
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chrX_-_106607352 | 1.69 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr9_+_65534704 | 1.67 |
ENSRNOT00000016730
|
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr4_-_2201749 | 1.61 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr9_-_43454078 | 1.61 |
ENSRNOT00000023550
|
Tmem131
|
transmembrane protein 131 |
| chr10_-_87459652 | 1.55 |
ENSRNOT00000018804
|
Krt40
|
keratin 40 |
| chr8_+_99568958 | 1.54 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr15_-_11912806 | 1.44 |
ENSRNOT00000068171
ENSRNOT00000008759 ENSRNOT00000049771 |
Slc4a7
|
solute carrier family 4 member 7 |
| chr3_+_131351587 | 1.40 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr1_-_93949187 | 1.30 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr15_-_40542495 | 1.29 |
ENSRNOT00000063943
|
Nup58
|
nucleoporin 58 |
| chr17_-_80860195 | 1.28 |
ENSRNOT00000038651
|
Trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr11_-_28842287 | 1.26 |
ENSRNOT00000061610
|
Krtap19-5
|
keratin associated protein 19-5 |
| chr2_-_9472384 | 1.24 |
ENSRNOT00000084295
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr8_+_17421557 | 1.24 |
ENSRNOT00000030569
|
Chordc1
|
cysteine and histidine rich domain containing 1 |
| chr3_-_77677349 | 1.23 |
ENSRNOT00000084989
|
Olr669
|
olfactory receptor 669 |
| chr18_-_40218225 | 1.21 |
ENSRNOT00000004723
|
Pggt1b
|
protein geranylgeranyltransferase type 1 subunit beta |
| chr10_-_92008082 | 1.17 |
ENSRNOT00000006361
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr10_-_88000423 | 1.13 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr20_-_6556350 | 1.10 |
ENSRNOT00000035819
|
Lemd2
|
LEM domain containing 2 |
| chr1_+_168378637 | 1.07 |
ENSRNOT00000028936
|
Olr86
|
olfactory receptor 86 |
| chr2_-_46042636 | 1.07 |
ENSRNOT00000087161
|
AABR07008152.1
|
|
| chr1_-_225077079 | 1.06 |
ENSRNOT00000074747
|
Uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr7_+_66717434 | 1.05 |
ENSRNOT00000077878
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr11_-_29710849 | 1.04 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr9_+_79659251 | 1.03 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr1_-_134867001 | 1.01 |
ENSRNOT00000090149
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr17_-_29895386 | 0.96 |
ENSRNOT00000048757
|
Cdyl
|
chromodomain Y-like |
| chr14_-_23002011 | 0.95 |
ENSRNOT00000002736
|
Ythdc1
|
YTH domain containing 1 |
| chr3_-_105663457 | 0.92 |
ENSRNOT00000067646
|
Zfp770
|
zinc finger protein 770 |
| chr8_+_67615635 | 0.90 |
ENSRNOT00000008971
|
Itga11
|
integrin subunit alpha 11 |
| chr6_+_69971227 | 0.89 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr7_-_143187594 | 0.88 |
ENSRNOT00000011732
ENSRNOT00000083466 |
Krt84
|
keratin 84 |
| chr7_+_107406962 | 0.78 |
ENSRNOT00000093254
|
Phf20l1
|
PHD finger protein 20-like protein 1 |
| chr1_+_154131926 | 0.73 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr5_-_16140896 | 0.72 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr1_+_261753364 | 0.70 |
ENSRNOT00000076659
|
R3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr7_-_3123297 | 0.56 |
ENSRNOT00000061878
|
Rab5b
|
RAB5B, member RAS oncogene family |
| chr3_+_76179214 | 0.55 |
ENSRNOT00000071281
|
LOC686677
|
similar to olfactory receptor 1161 |
| chr2_-_173800761 | 0.54 |
ENSRNOT00000039524
|
Wdr49
|
WD repeat domain 49 |
| chrX_+_20216587 | 0.50 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr18_-_5314511 | 0.42 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr9_+_88494676 | 0.41 |
ENSRNOT00000089451
|
LOC102556337
|
mitochondrial fission factor-like |
| chr10_-_61232331 | 0.41 |
ENSRNOT00000065045
|
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr17_+_70262363 | 0.37 |
ENSRNOT00000048933
|
Fam208b
|
family with sequence similarity 208, member B |
| chr4_-_34194764 | 0.37 |
ENSRNOT00000045270
|
Col28a1
|
collagen type XXVIII alpha 1 chain |
| chr14_+_17210733 | 0.36 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr7_-_63045728 | 0.27 |
ENSRNOT00000039532
|
Lemd3
|
LEM domain containing 3 |
| chr8_+_115546712 | 0.26 |
ENSRNOT00000079801
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr2_-_43067956 | 0.25 |
ENSRNOT00000090616
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr9_-_93125014 | 0.22 |
ENSRNOT00000023829
|
Htr2b
|
5-hydroxytryptamine receptor 2B |
| chr16_-_37177033 | 0.15 |
ENSRNOT00000014015
|
Fbxo8
|
F-box protein 8 |
| chr20_+_11655967 | 0.12 |
ENSRNOT00000064565
|
LOC690478
|
similar to keratin associated protein 10-7 |
| chr16_-_85464002 | 0.10 |
ENSRNOT00000037018
|
AABR07026596.1
|
|
| chr19_-_19896421 | 0.09 |
ENSRNOT00000020793
|
Heatr3
|
HEAT repeat containing 3 |
| chr3_+_46185360 | 0.06 |
ENSRNOT00000051344
|
LOC100361645
|
LRRGT00075-like |
| chr10_+_38918748 | 0.05 |
ENSRNOT00000009999
|
Kif3a
|
kinesin family member 3a |
| chr11_-_28614260 | 0.05 |
ENSRNOT00000074102
|
Krtap26-1
|
keratin associated protein 26-1 |
| chr5_+_127489418 | 0.01 |
ENSRNOT00000065275
|
RGD1559786
|
similar to RIKEN cDNA 0610037L13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc13_Hoxd13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 23.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 2.6 | 12.9 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 2.4 | 7.3 | GO:0080033 | cellular response to carbon monoxide(GO:0071245) cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 2.4 | 12.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 1.5 | 6.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.1 | 3.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 1.0 | 3.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.9 | 9.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.9 | 2.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.8 | 5.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.8 | 7.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.7 | 2.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.6 | 8.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 3.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.4 | 1.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 | 5.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 2.5 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.3 | 1.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.3 | 1.4 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.3 | 1.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.2 | 1.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 3.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.2 | 1.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 3.6 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.2 | 6.2 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 1.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 5.1 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 1.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 1.4 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 12.9 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.1 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 15.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 4.7 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 | 0.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 1.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.9 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.4 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.0 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 5.3 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.1 | GO:2000769 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.0 | 2.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 2.4 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 1.6 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 2.2 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 3.6 | GO:0045137 | development of primary sexual characteristics(GO:0045137) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.6 | 2.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.5 | 3.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.5 | 3.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 12.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 1.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.3 | 19.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 1.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 3.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.2 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.2 | 2.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 3.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 7.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 14.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 7.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.8 | 9.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.1 | 5.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.8 | 3.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.7 | 3.4 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.6 | 2.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 5.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 12.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 3.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 2.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 14.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 6.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.3 | 1.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 1.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.3 | 1.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.3 | 3.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 3.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 8.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 3.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 12.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 0.9 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 3.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 7.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 4.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 4.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 5.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 19.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 5.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 5.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 8.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 12.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 9.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 1.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 5.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 15.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 3.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 4.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.6 | 8.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.5 | 12.9 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.4 | 5.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 3.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 7.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 12.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.2 | 4.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 11.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 5.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 14.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 5.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |