Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
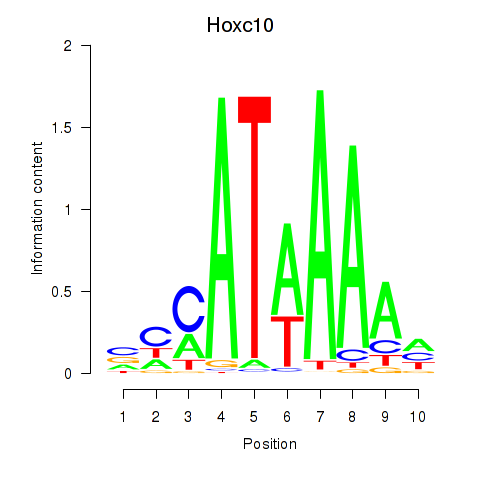
Results for Hoxc10
Z-value: 0.43
Transcription factors associated with Hoxc10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc10
|
ENSRNOG00000016149 | homeo box C10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc10 | rn6_v1_chr7_+_144577465_144577465 | -0.09 | 1.0e-01 | Click! |
Activity profile of Hoxc10 motif
Sorted Z-values of Hoxc10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_77485113 | 12.44 |
ENSRNOT00000080254
|
Tnr
|
tenascin R |
| chr5_+_117698764 | 10.94 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr2_-_154418920 | 9.95 |
ENSRNOT00000076326
|
Plch1
|
phospholipase C, eta 1 |
| chr15_-_14737704 | 9.33 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr13_+_42220251 | 9.22 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr14_-_20920286 | 8.89 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr2_-_154418629 | 8.74 |
ENSRNOT00000076274
ENSRNOT00000076165 |
Plch1
|
phospholipase C, eta 1 |
| chr13_+_44424689 | 7.99 |
ENSRNOT00000005206
|
Acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr1_-_211923929 | 7.65 |
ENSRNOT00000054887
|
Nkx6-2
|
NK6 homeobox 2 |
| chr3_+_159902441 | 7.48 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr15_-_23969011 | 7.41 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr1_-_43638161 | 6.34 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_69549673 | 6.19 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr5_+_167141875 | 6.04 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr12_-_35979193 | 5.68 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr10_-_66848388 | 5.04 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr7_-_140437467 | 4.94 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr20_-_9876008 | 4.89 |
ENSRNOT00000001537
|
Tff2
|
trefoil factor 2 |
| chr3_-_141411170 | 4.86 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr10_-_40953467 | 4.64 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chr5_+_167142182 | 4.61 |
ENSRNOT00000024054
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr20_-_54517709 | 4.25 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr7_-_143420027 | 3.90 |
ENSRNOT00000082863
|
Krt2
|
keratin 2 |
| chr1_-_89369960 | 3.20 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr10_-_87954055 | 3.12 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr10_-_71382058 | 3.11 |
ENSRNOT00000043148
|
Dusp14
|
dual specificity phosphatase 14 |
| chr3_+_59981959 | 2.96 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr3_+_21114856 | 2.77 |
ENSRNOT00000044947
|
Olr423
|
olfactory receptor 423 |
| chr19_-_14945302 | 2.71 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr7_-_143228060 | 2.67 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr8_-_93390305 | 2.59 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr11_-_28614260 | 2.51 |
ENSRNOT00000074102
|
Krtap26-1
|
keratin associated protein 26-1 |
| chr3_-_105663457 | 2.49 |
ENSRNOT00000067646
|
Zfp770
|
zinc finger protein 770 |
| chr11_+_80255790 | 2.47 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr1_-_134867001 | 2.33 |
ENSRNOT00000090149
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_-_87962846 | 2.25 |
ENSRNOT00000018176
|
Krt31
|
keratin 31 |
| chr20_-_5064469 | 2.22 |
ENSRNOT00000001120
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr17_-_43689311 | 2.19 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chrX_-_9999401 | 1.88 |
ENSRNOT00000060992
|
Gpr82
|
G protein-coupled receptor 82 |
| chr10_+_56411028 | 1.71 |
ENSRNOT00000085772
|
AABR07029862.1
|
|
| chr9_+_66058047 | 1.66 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr3_+_137154086 | 1.61 |
ENSRNOT00000034252
|
Otor
|
otoraplin |
| chr18_+_29966245 | 1.55 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr6_-_38007162 | 1.52 |
ENSRNOT00000044583
|
AABR07063632.1
|
|
| chr1_+_70437776 | 1.50 |
ENSRNOT00000020448
|
Olr3
|
olfactory receptor 3 |
| chr5_-_153840178 | 1.46 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr10_+_10761477 | 1.36 |
ENSRNOT00000004176
|
Rogdi
|
rogdi homolog |
| chr10_+_84031955 | 1.27 |
ENSRNOT00000009934
|
Hoxb13
|
homeo box B13 |
| chr1_+_260798239 | 1.18 |
ENSRNOT00000036791
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr10_-_87500919 | 1.09 |
ENSRNOT00000084798
|
Krtap1-5
|
keratin associated protein 1-5 |
| chr7_-_143167772 | 1.09 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr10_-_87153982 | 1.06 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr9_+_20048121 | 0.99 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr14_-_105069565 | 0.98 |
ENSRNOT00000067365
|
AABR07016578.1
|
|
| chr3_-_79121620 | 0.97 |
ENSRNOT00000087141
|
Olr741
|
olfactory receptor 741 |
| chr3_+_76179214 | 0.76 |
ENSRNOT00000071281
|
LOC686677
|
similar to olfactory receptor 1161 |
| chr7_-_143187594 | 0.76 |
ENSRNOT00000011732
ENSRNOT00000083466 |
Krt84
|
keratin 84 |
| chr16_-_85464002 | 0.67 |
ENSRNOT00000037018
|
AABR07026596.1
|
|
| chr13_+_26463404 | 0.56 |
ENSRNOT00000061779
|
LOC103690041
|
serpin B12-like |
| chr19_-_34752695 | 0.54 |
ENSRNOT00000052018
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr1_+_8310577 | 0.54 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr3_+_67538289 | 0.48 |
ENSRNOT00000009839
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr19_-_22194740 | 0.41 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_+_99616447 | 0.40 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr18_+_29124428 | 0.27 |
ENSRNOT00000073131
|
Igip
|
IgA-inducing protein |
| chr16_+_70074672 | 0.18 |
ENSRNOT00000073167
ENSRNOT00000035638 |
AABR07026271.1
|
|
| chr15_-_34693034 | 0.17 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr3_-_76135475 | 0.13 |
ENSRNOT00000007712
|
Olr604
|
olfactory receptor 604 |
| chr10_-_87484935 | 0.05 |
ENSRNOT00000016752
|
Krtap3-1
|
keratin associated protein 3-1 |
| chr2_-_178521038 | 0.04 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 2.2 | 10.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 2.1 | 10.7 | GO:0015755 | fructose transport(GO:0015755) |
| 2.1 | 12.4 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.9 | 7.5 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 1.6 | 4.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.5 | 7.6 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 1.3 | 8.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.8 | 3.2 | GO:0034760 | negative regulation of iron ion transmembrane transport(GO:0034760) cellular response to bile acid(GO:1903413) |
| 0.7 | 4.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.6 | 2.5 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.6 | 4.6 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.3 | 8.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 3.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 4.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.5 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.2 | 5.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.1 | 2.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 4.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.5 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 18.7 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 2.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 2.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 1.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.4 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 2.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.0 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 2.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.7 | 4.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.4 | 9.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 3.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 5.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 5.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 9.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 10.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 8.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 7.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 8.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.6 | GO:0005657 | replication fork(GO:0005657) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.7 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.5 | 4.6 | GO:0030977 | taurine binding(GO:0030977) |
| 1.2 | 10.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 1.2 | 10.7 | GO:0070061 | fructose binding(GO:0070061) |
| 1.1 | 7.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.7 | 3.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.6 | 7.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.6 | 4.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 8.9 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.3 | 4.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 2.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 2.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 8.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 2.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 2.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 4.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 4.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 12.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 3.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 4.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0051787 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) misfolded protein binding(GO:0051787) |
| 0.0 | 2.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 6.5 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 7.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 12.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 10.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 10.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.6 | 7.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.5 | 12.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 4.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 8.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 5.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |