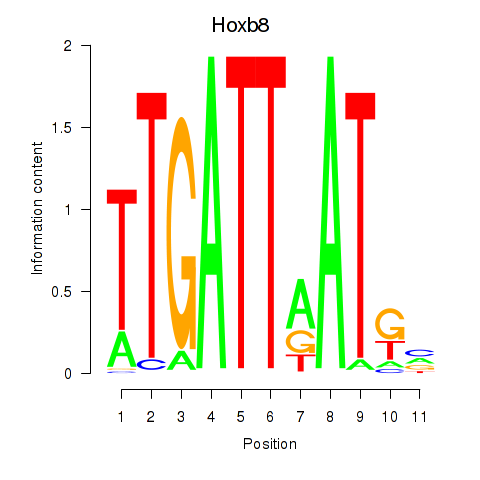
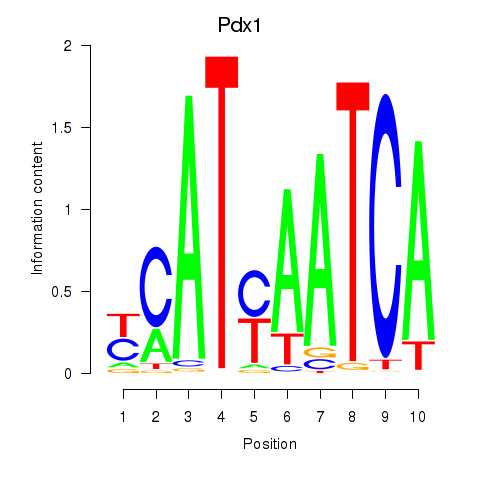
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxb8_Pdx1
Z-value: 1.06
Transcription factors associated with Hoxb8_Pdx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb8
|
ENSRNOG00000007585 | homeo box B8 |
|
Pdx1
|
ENSRNOG00000046458 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb8 | rn6_v1_chr10_+_84131495_84131495 | 0.33 | 1.5e-09 | Click! |
| Pdx1 | rn6_v1_chr12_-_9501213_9501213 | 0.11 | 5.4e-02 | Click! |
Activity profile of Hoxb8_Pdx1 motif
Sorted Z-values of Hoxb8_Pdx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_201620642 | 64.51 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr2_-_105089659 | 56.60 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr7_+_125288081 | 54.49 |
ENSRNOT00000085216
|
Parvg
|
parvin, gamma |
| chr16_-_29936307 | 44.44 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr3_-_23020441 | 38.09 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr1_-_197821936 | 33.68 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr10_+_84200880 | 29.37 |
ENSRNOT00000011213
|
Hoxb2
|
homeobox B2 |
| chr16_-_31301880 | 25.24 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr16_-_71040847 | 25.16 |
ENSRNOT00000020606
|
Star
|
steroidogenic acute regulatory protein |
| chr4_-_82133605 | 23.49 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr9_-_42839837 | 22.96 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr7_+_37812831 | 21.73 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr8_-_114617466 | 16.96 |
ENSRNOT00000082992
ENSRNOT00000038160 |
Col6a5
|
collagen type VI alpha 5 chain |
| chr6_-_143195143 | 16.59 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chr2_-_200762492 | 15.61 |
ENSRNOT00000056172
|
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr2_-_200762206 | 15.56 |
ENSRNOT00000068511
ENSRNOT00000086835 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr5_-_2803855 | 15.40 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr9_-_52238564 | 15.17 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr5_-_12172009 | 14.49 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr9_-_14706557 | 14.34 |
ENSRNOT00000048975
|
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr1_+_15834779 | 14.24 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr1_+_264893162 | 13.71 |
ENSRNOT00000021714
|
Tlx1
|
T-cell leukemia, homeobox 1 |
| chr2_-_203680083 | 13.62 |
ENSRNOT00000021268
|
Cd2
|
Cd2 molecule |
| chr9_+_112360419 | 13.16 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr3_+_19495628 | 13.01 |
ENSRNOT00000077990
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr11_+_53140599 | 12.78 |
ENSRNOT00000083438
|
Bbx
|
BBX, HMG-box containing |
| chrX_+_119390013 | 12.58 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr8_-_77398156 | 12.50 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr8_+_44136496 | 12.47 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr15_-_36798814 | 12.21 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr17_-_39824299 | 11.70 |
ENSRNOT00000023412
|
Prl
|
prolactin |
| chr3_+_148579920 | 11.54 |
ENSRNOT00000012432
|
Hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr1_-_16687817 | 11.48 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr13_-_83457888 | 11.42 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chrX_-_123486814 | 11.05 |
ENSRNOT00000016993
|
RGD1564541
|
similar to hypothetical protein FLJ22965 |
| chr1_-_198476476 | 10.94 |
ENSRNOT00000027525
|
Kif22
|
kinesin family member 22 |
| chr5_-_76261198 | 10.83 |
ENSRNOT00000087380
|
AABR07048387.1
|
|
| chr13_+_27465930 | 10.82 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr20_-_5037022 | 10.80 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr16_-_79700992 | 10.23 |
ENSRNOT00000016420
|
Kbtbd11
|
kelch repeat and BTB domain containing 11 |
| chr2_+_235596907 | 10.10 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr5_-_50638136 | 10.08 |
ENSRNOT00000012519
|
Mob3b
|
MOB kinase activator 3B |
| chr1_-_166912524 | 10.07 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr4_+_57952982 | 10.07 |
ENSRNOT00000014465
|
Cpa1
|
carboxypeptidase A1 |
| chrX_+_35869538 | 9.98 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr3_-_55451798 | 9.93 |
ENSRNOT00000008837
|
Spc25
|
SPC25, NDC80 kinetochore complex component |
| chr4_-_168517177 | 9.89 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr11_+_85508300 | 9.83 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr2_+_212247451 | 9.68 |
ENSRNOT00000027813
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr3_+_17546566 | 9.66 |
ENSRNOT00000050825
|
AABR07051583.1
|
|
| chr2_-_91497091 | 9.45 |
ENSRNOT00000015185
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr9_-_119869639 | 9.40 |
ENSRNOT00000080498
|
Ndc80
|
NDC80 kinetochore complex component |
| chr6_-_140407307 | 9.28 |
ENSRNOT00000079087
|
AABR07065768.3
|
|
| chr4_-_163849618 | 9.24 |
ENSRNOT00000086363
ENSRNOT00000077637 |
Ly49si1
|
immunoreceptor Ly49si1 |
| chr11_+_85532526 | 9.15 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr4_+_70689737 | 9.10 |
ENSRNOT00000018852
|
Prss2
|
protease, serine, 2 |
| chr6_+_132510757 | 8.94 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr4_+_102876038 | 8.81 |
ENSRNOT00000085205
|
AABR07061022.3
|
|
| chr11_+_85618714 | 8.73 |
ENSRNOT00000074614
|
AC109901.1
|
|
| chr2_+_55835151 | 8.68 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr8_-_104155775 | 8.60 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr14_+_43810119 | 8.55 |
ENSRNOT00000003327
|
Rbm47
|
RNA binding motif protein 47 |
| chr15_-_33285779 | 8.52 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chr1_-_102826965 | 8.38 |
ENSRNOT00000078692
|
Saa4
|
serum amyloid A4 |
| chr9_+_8093262 | 8.36 |
ENSRNOT00000080314
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr3_+_145764932 | 8.34 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr7_+_11660934 | 8.29 |
ENSRNOT00000022336
|
Lmnb2
|
lamin B2 |
| chr10_+_61685645 | 8.26 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr13_-_61591139 | 8.09 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr4_+_55720010 | 7.94 |
ENSRNOT00000063987
|
Fscn3
|
fascin actin-bundling protein 3 |
| chr11_+_30904733 | 7.90 |
ENSRNOT00000036027
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr14_+_45062662 | 7.90 |
ENSRNOT00000059124
|
Tlr10
|
toll-like receptor 10 |
| chr8_+_132828091 | 7.83 |
ENSRNOT00000008269
|
Ccr9
|
C-C motif chemokine receptor 9 |
| chr20_-_4817146 | 7.70 |
ENSRNOT00000080174
|
Ddx39b
|
DExD-box helicase 39B |
| chr3_-_20479999 | 7.46 |
ENSRNOT00000050573
|
AABR07051733.2
|
|
| chr3_+_72395218 | 7.42 |
ENSRNOT00000057616
|
Prg3
|
proteoglycan 3, pro eosinophil major basic protein 2 |
| chr14_-_14390699 | 7.38 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr1_-_227932603 | 7.38 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr4_-_62526724 | 7.29 |
ENSRNOT00000038950
|
RGD1565367
|
similar to Solute carrier family 23, member 2 (Sodium-dependent vitamin C transporter 2) |
| chr4_-_129515435 | 7.08 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chrX_-_1952111 | 6.99 |
ENSRNOT00000061140
|
Jade3
|
jade family PHD finger 3 |
| chr1_-_189182306 | 6.96 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr10_-_75171774 | 6.90 |
ENSRNOT00000011735
|
Epx
|
eosinophil peroxidase |
| chr7_-_71139267 | 6.83 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr4_+_99823252 | 6.82 |
ENSRNOT00000013587
|
Polr1a
|
RNA polymerase I subunit A |
| chr3_-_72171078 | 6.81 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr1_+_219390523 | 6.81 |
ENSRNOT00000054852
|
Gpr152
|
G protein-coupled receptor 152 |
| chr6_+_139551751 | 6.77 |
ENSRNOT00000081684
|
AABR07065699.2
|
|
| chr7_+_133856101 | 6.76 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr14_+_3506339 | 6.72 |
ENSRNOT00000002867
|
Tgfbr3
|
transforming growth factor beta receptor 3 |
| chr4_+_88119838 | 6.70 |
ENSRNOT00000073173
|
Vom1r82
|
vomeronasal 1 receptor 82 |
| chr6_-_139041654 | 6.55 |
ENSRNOT00000075664
|
AABR07065656.1
|
|
| chrX_+_984798 | 6.51 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr6_+_106052212 | 6.48 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_+_140601791 | 6.46 |
ENSRNOT00000091588
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr4_-_167202106 | 6.38 |
ENSRNOT00000038581
|
Tas2r140
|
taste receptor, type 2, member 140 |
| chr11_+_85561460 | 6.36 |
ENSRNOT00000075455
|
AABR07072262.1
|
|
| chr2_-_41871858 | 6.35 |
ENSRNOT00000039720
|
Gapt
|
Grb2-binding adaptor protein, transmembrane |
| chr10_+_47930633 | 6.28 |
ENSRNOT00000003515
|
Grap
|
GRB2-related adaptor protein |
| chr9_+_67774150 | 6.27 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr1_+_84280945 | 6.26 |
ENSRNOT00000057190
|
Sertad3
|
SERTA domain containing 3 |
| chr9_+_79944132 | 6.22 |
ENSRNOT00000071856
ENSRNOT00000022459 |
Smarcal1
|
Swi/SNF related matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr6_-_138640187 | 6.18 |
ENSRNOT00000087983
|
AABR07065651.6
|
|
| chr6_+_139209936 | 6.10 |
ENSRNOT00000087620
|
AABR07065680.1
|
|
| chr3_+_61658245 | 6.03 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr4_-_162856663 | 6.01 |
ENSRNOT00000077791
|
Clec2e
|
C-type lectin domain family 2, member E |
| chr10_-_89454681 | 5.96 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr1_-_124803363 | 5.96 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr1_-_189181901 | 5.95 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr15_-_34693034 | 5.93 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr6_-_139747737 | 5.92 |
ENSRNOT00000090626
|
AABR07065705.3
|
|
| chr20_+_8312792 | 5.83 |
ENSRNOT00000083108
|
Cmtr1
|
cap methyltransferase 1 |
| chr17_-_42276797 | 5.83 |
ENSRNOT00000083996
|
LOC100911664
|
uncharacterized LOC100911664 |
| chr11_+_17538063 | 5.79 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr17_+_9596957 | 5.79 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr1_-_271256379 | 5.74 |
ENSRNOT00000081605
|
LOC100912218
|
inositol 1,4,5-trisphosphate receptor-interacting protein-like |
| chr1_-_267653389 | 5.69 |
ENSRNOT00000082550
|
LOC100912218
|
inositol 1,4,5-trisphosphate receptor-interacting protein-like |
| chr3_-_118635268 | 5.68 |
ENSRNOT00000086357
|
Atp8b4
|
ATPase phospholipid transporting 8B4 (putative) |
| chr3_-_47025128 | 5.66 |
ENSRNOT00000011682
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr5_-_164348713 | 5.62 |
ENSRNOT00000087044
|
LOC500584
|
similar to casein kinase 1, gamma 3 isoform 2 |
| chr16_-_11214948 | 5.40 |
ENSRNOT00000023369
|
LOC690468
|
similar to 60S ribosomal protein L38 |
| chr9_-_88748866 | 5.40 |
ENSRNOT00000071906
|
Rpl30l1
|
ribosomal protein L30-like 1 |
| chr3_+_118317761 | 5.40 |
ENSRNOT00000012700
|
Fgf7
|
fibroblast growth factor 7 |
| chr4_-_164536556 | 5.39 |
ENSRNOT00000087796
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr6_-_138536162 | 5.39 |
ENSRNOT00000083031
|
AABR07065643.1
|
|
| chr12_-_38274036 | 5.32 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr16_+_11032531 | 5.31 |
ENSRNOT00000078390
|
Wapl
|
WAPL cohesin release factor |
| chr20_-_5166252 | 5.28 |
ENSRNOT00000001138
|
Aif1
|
allograft inflammatory factor 1 |
| chr16_+_61926586 | 5.28 |
ENSRNOT00000058884
|
AABR07026059.1
|
|
| chr6_+_139345486 | 5.22 |
ENSRNOT00000081540
|
AABR07065688.1
|
|
| chr6_-_138948583 | 5.20 |
ENSRNOT00000084614
|
AABR07065656.2
|
|
| chr1_+_7726529 | 5.12 |
ENSRNOT00000073460
|
Adat2
|
adenosine deaminase, tRNA-specific 2 |
| chr7_+_63467216 | 5.10 |
ENSRNOT00000064349
|
LOC100909505
|
N-acetylglucosamine-6-sulfatase-like |
| chr4_-_165456677 | 4.93 |
ENSRNOT00000082207
|
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr6_-_141008427 | 4.89 |
ENSRNOT00000074472
|
AABR07065778.2
|
|
| chr9_+_67699379 | 4.86 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr17_-_2660902 | 4.85 |
ENSRNOT00000067979
|
Mfsd14b
|
major facilitator superfamily domain containing 14B |
| chr18_+_79773608 | 4.84 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr6_+_29106141 | 4.84 |
ENSRNOT00000084560
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr1_+_199495298 | 4.83 |
ENSRNOT00000086003
ENSRNOT00000026748 |
Itgad
|
integrin subunit alpha D |
| chr6_-_138679665 | 4.79 |
ENSRNOT00000086777
|
AABR07065651.4
|
|
| chr20_+_42966140 | 4.78 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chrX_+_112311251 | 4.72 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr13_-_82295123 | 4.72 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr4_+_14039977 | 4.71 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr12_-_52396066 | 4.70 |
ENSRNOT00000056712
|
Lrcol1
|
leucine rich colipase-like 1 |
| chr16_-_23991570 | 4.70 |
ENSRNOT00000044198
ENSRNOT00000018854 |
Nat2
Nat1
|
N-acetyltransferase 2 N-acetyltransferase 1 |
| chr4_-_170740274 | 4.58 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr1_-_101095594 | 4.58 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr16_-_7412150 | 4.55 |
ENSRNOT00000047252
|
RGD1564325
|
similar to ribosomal protein S24 |
| chr4_-_132740938 | 4.52 |
ENSRNOT00000007185
|
Rybp
|
RING1 and YY1 binding protein |
| chr17_+_23135985 | 4.51 |
ENSRNOT00000090794
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr6_+_135610743 | 4.50 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr6_-_138948424 | 4.49 |
ENSRNOT00000072566
|
AABR07065656.2
|
|
| chr7_-_1738642 | 4.38 |
ENSRNOT00000075554
|
AABR07055356.1
|
|
| chr11_-_60931794 | 4.36 |
ENSRNOT00000018473
|
Nepro
|
nucleolus and neural progenitor protein |
| chr16_+_2634603 | 4.34 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr1_-_48797631 | 4.31 |
ENSRNOT00000082958
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr4_-_82177420 | 4.30 |
ENSRNOT00000074452
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr17_-_90522091 | 4.30 |
ENSRNOT00000077767
|
Lyst
|
lysosomal trafficking regulator |
| chrX_+_21499934 | 4.28 |
ENSRNOT00000090890
ENSRNOT00000081037 |
Huwe1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr14_+_19319299 | 4.27 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr11_+_88376256 | 4.26 |
ENSRNOT00000081776
|
Vpreb1
|
pre-B lymphocyte 1 |
| chr4_-_162230859 | 4.25 |
ENSRNOT00000042872
|
LOC689757
|
similar to osteoclast inhibitory lectin |
| chr8_-_92942076 | 4.23 |
ENSRNOT00000056937
|
Fam46a
|
family with sequence similarity 46, member A |
| chr4_-_131694755 | 4.22 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr15_+_60084918 | 4.22 |
ENSRNOT00000012632
|
Epsti1
|
epithelial stromal interaction 1 |
| chr1_+_22332090 | 4.20 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr6_-_138679506 | 4.16 |
ENSRNOT00000075376
|
AABR07065651.4
|
|
| chr5_+_44270057 | 4.11 |
ENSRNOT00000042939
|
RGD1562265
|
similar to ribosomal protein S12 |
| chr8_-_52828134 | 4.10 |
ENSRNOT00000007736
|
Rbm7
|
RNA binding motif protein 7 |
| chr15_+_4254499 | 4.08 |
ENSRNOT00000061686
|
Anxa7
|
annexin A7 |
| chr7_-_14189688 | 4.06 |
ENSRNOT00000037456
|
Notch3
|
notch 3 |
| chr5_+_48303366 | 4.03 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chrX_+_124894466 | 4.00 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr13_-_68253251 | 3.98 |
ENSRNOT00000076776
|
Hmcn1
|
hemicentin 1 |
| chrX_+_77119911 | 3.96 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chr14_-_72122158 | 3.91 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr2_-_196526886 | 3.88 |
ENSRNOT00000077325
|
Setdb1
|
SET domain, bifurcated 1 |
| chr4_+_166601960 | 3.87 |
ENSRNOT00000073394
|
AABR07062266.1
|
|
| chr11_-_29638922 | 3.83 |
ENSRNOT00000048888
|
LOC100359671
|
ribosomal protein L29-like |
| chr13_-_83917001 | 3.81 |
ENSRNOT00000004387
ENSRNOT00000082422 |
Rcsd1
|
RCSD domain containing 1 |
| chr5_+_135412256 | 3.81 |
ENSRNOT00000076094
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr8_+_99632803 | 3.80 |
ENSRNOT00000087190
|
Plscr1
|
phospholipid scramblase 1 |
| chr14_+_1937041 | 3.78 |
ENSRNOT00000000040
|
Tmed11
|
transmembrane emp24 protein transport domain containing 11 |
| chr16_-_21338771 | 3.78 |
ENSRNOT00000014265
|
Pbx4
|
PBX homeobox 4 |
| chr4_-_82263302 | 3.77 |
ENSRNOT00000082933
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr10_-_87153982 | 3.75 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr3_+_4867233 | 3.73 |
ENSRNOT00000006754
|
Rpl7a
|
ribosomal protein L7a |
| chrX_-_1856258 | 3.72 |
ENSRNOT00000079980
|
RGD1564855
|
similar to RuvB-like protein 1 |
| chr5_+_117586103 | 3.64 |
ENSRNOT00000084640
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr15_+_4400991 | 3.64 |
ENSRNOT00000008553
|
Ecd
|
ecdysoneless cell cycle regulator |
| chr13_-_83202864 | 3.55 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr6_-_139654508 | 3.49 |
ENSRNOT00000082576
|
AABR07065705.5
|
|
| chr18_+_27632786 | 3.42 |
ENSRNOT00000073564
ENSRNOT00000078969 |
Reep2
|
receptor accessory protein 2 |
| chr5_-_150994913 | 3.41 |
ENSRNOT00000075506
|
RGD1561465
|
similar to RIKEN cDNA 2900010J23 |
| chr4_-_82263117 | 3.40 |
ENSRNOT00000008542
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr17_-_4123302 | 3.39 |
ENSRNOT00000039754
|
RGD1564827
|
similar to cathepsin M |
| chr5_+_47546014 | 3.39 |
ENSRNOT00000065882
|
Bach2
|
BTB domain and CNC homolog 2 |
| chr7_+_2300434 | 3.36 |
ENSRNOT00000071785
|
AABR07055413.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb8_Pdx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 29.4 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 4 development(GO:0021570) |
| 9.5 | 38.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 8.4 | 25.2 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 5.9 | 35.1 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 4.2 | 12.6 | GO:0035566 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) regulation of metanephros size(GO:0035566) |
| 4.2 | 12.5 | GO:0010034 | response to acetate(GO:0010034) |
| 3.8 | 11.5 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 3.8 | 15.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 3.6 | 71.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 3.6 | 3.6 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 3.2 | 12.9 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 3.1 | 12.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 2.5 | 35.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 2.5 | 7.5 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 2.5 | 7.4 | GO:0045575 | basophil activation(GO:0045575) |
| 2.4 | 14.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 2.4 | 9.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 2.3 | 6.9 | GO:0002215 | defense response to nematode(GO:0002215) |
| 2.2 | 6.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 2.1 | 4.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 2.1 | 8.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 2.0 | 6.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.0 | 21.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.9 | 5.8 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 1.8 | 7.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.8 | 5.3 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.8 | 5.3 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 1.6 | 6.4 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.6 | 3.2 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 1.5 | 6.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.5 | 11.7 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.4 | 4.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 1.2 | 4.9 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 1.2 | 13.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 1.2 | 9.4 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.1 | 13.6 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.1 | 7.7 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 1.0 | 4.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 1.0 | 2.9 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 1.0 | 6.8 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 1.0 | 2.9 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 0.9 | 2.8 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.9 | 10.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.9 | 10.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.9 | 10.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.8 | 2.5 | GO:0016488 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.8 | 2.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.8 | 4.7 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.8 | 7.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.8 | 2.3 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.7 | 3.7 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.7 | 8.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.7 | 4.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.7 | 4.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.6 | 3.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.6 | 2.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.6 | 1.8 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.6 | 11.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.6 | 58.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.6 | 2.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.6 | 2.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.6 | 7.5 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.6 | 7.4 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.6 | 6.8 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.6 | 6.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.5 | 3.8 | GO:2000371 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.5 | 6.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.5 | 2.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 4.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.5 | 3.2 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.5 | 1.6 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.5 | 4.1 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.5 | 2.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 | 4.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.5 | 1.5 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.5 | 3.5 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.5 | 12.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.5 | 4.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.5 | 1.9 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.5 | 4.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 7.8 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.4 | 2.6 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 | 3.4 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.4 | 10.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.4 | 12.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.4 | 1.2 | GO:0033986 | response to methanol(GO:0033986) |
| 0.4 | 2.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 9.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.4 | 1.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 | 1.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.4 | 1.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.4 | 2.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.4 | 15.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.4 | 2.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 10.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.3 | 1.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 4.7 | GO:0097068 | response to thyroxine(GO:0097068) |
| 0.3 | 2.7 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.3 | 6.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.3 | 4.6 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.3 | 2.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.3 | 13.7 | GO:0048536 | spleen development(GO:0048536) |
| 0.3 | 3.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 4.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.3 | 11.4 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.3 | 1.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.3 | 5.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 3.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.3 | 12.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.3 | 3.8 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.2 | 9.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 1.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 2.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 2.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 1.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.8 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 1.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 2.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 3.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 2.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.3 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.2 | 4.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 8.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 4.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 2.5 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.1 | 7.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 2.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 9.3 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.1 | 2.0 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 1.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 8.3 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 4.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 2.6 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.0 | GO:0035745 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 3.2 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 1.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 2.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.9 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 1.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 6.2 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 3.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 2.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.6 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.1 | 1.6 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 4.2 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.1 | 1.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 5.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 4.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 2.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 1.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.4 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.1 | 0.7 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 3.6 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 | 1.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 1.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 6.1 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 4.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 2.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.4 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 2.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 4.6 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.6 | GO:0032438 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 4.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 2.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 3.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 2.1 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.6 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 3.8 | 15.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.4 | 64.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 2.2 | 6.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.8 | 5.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.5 | 6.0 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 1.3 | 21.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.2 | 4.8 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.9 | 2.8 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.9 | 2.6 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.9 | 7.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.7 | 2.9 | GO:0044299 | C-fiber(GO:0044299) |
| 0.7 | 2.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.7 | 20.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.7 | 4.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.6 | 7.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.6 | 5.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.6 | 9.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.6 | 3.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.6 | 6.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.6 | 3.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.5 | 2.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 2.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.5 | 1.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 2.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.4 | 2.5 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.4 | 6.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 5.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 3.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 16.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 3.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 37.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.3 | 1.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 0.8 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 9.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 16.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 12.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 3.8 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 2.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 5.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.9 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 10.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 14.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 1.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 4.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 8.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 14.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 2.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 10.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 5.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 8.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 9.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 6.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 2.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 27.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 4.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 7.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 25.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 4.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 4.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 3.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 6.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 2.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 3.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 10.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 8.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 2.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 6.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 33.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 2.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 9.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 31.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 9.1 | 63.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 3.6 | 14.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 2.6 | 7.9 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 2.5 | 12.6 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 2.4 | 64.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 2.3 | 11.5 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 2.1 | 12.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.9 | 5.8 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 1.8 | 11.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.6 | 4.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.5 | 7.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.4 | 7.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.3 | 6.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 1.1 | 10.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 1.1 | 25.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 1.1 | 5.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.0 | 9.4 | GO:0043559 | insulin binding(GO:0043559) |
| 1.0 | 13.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 1.0 | 7.7 | GO:0030621 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.9 | 4.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.9 | 2.8 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.9 | 2.8 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.9 | 2.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.9 | 4.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.9 | 2.7 | GO:0008254 | 3'-nucleotidase activity(GO:0008254) |
| 0.8 | 10.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.8 | 2.5 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.8 | 2.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.8 | 4.0 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 0.8 | 6.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.7 | 9.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.7 | 2.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.7 | 6.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.7 | 6.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.7 | 8.5 | GO:0070097 | alpha-catenin binding(GO:0045294) delta-catenin binding(GO:0070097) |
| 0.6 | 1.9 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.6 | 2.5 | GO:0001635 | calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.6 | 1.8 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.6 | 2.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.6 | 2.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.5 | 6.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.5 | 3.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 7.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.5 | 4.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.5 | 2.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 1.4 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.4 | 39.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.4 | 3.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 3.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 2.5 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.4 | 6.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 2.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 1.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.4 | 4.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 6.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 10.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 2.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 3.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 1.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 3.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 4.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 6.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 26.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 11.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 6.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 4.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 1.0 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.3 | 5.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 3.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 2.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 0.9 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 2.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 7.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 6.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 4.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 30.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 4.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 9.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 13.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.2 | 2.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 2.0 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 6.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.0 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 2.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 2.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0030283 | testosterone dehydrogenase [NAD(P)] activity(GO:0030283) |
| 0.1 | 3.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 30.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 4.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 3.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 6.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.4 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.5 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 5.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 7.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 19.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 8.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 12.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 14.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 6.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 10.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 36.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 6.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 4.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 18.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 8.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 2.1 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 15.5 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.1 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 1.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 57.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.8 | 34.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.8 | 19.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.7 | 24.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.5 | 25.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 11.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.3 | 6.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 7.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 2.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 48.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 7.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 7.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 4.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 8.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 4.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 4.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 10.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 11.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 6.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 8.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 31.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 12.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 31.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 1.3 | 11.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 1.2 | 25.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 1.2 | 9.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.9 | 8.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.8 | 32.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.7 | 12.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 15.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.6 | 13.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.6 | 11.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.5 | 4.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 3.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.5 | 4.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 9.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.4 | 4.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 9.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.4 | 27.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.4 | 3.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 10.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 6.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 6.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 5.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.3 | 2.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 26.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 7.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 4.9 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.3 | 3.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 6.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.3 | 11.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 16.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 11.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 10.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 2.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 4.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 2.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 2.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 5.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 1.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.2 | 2.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 8.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 4.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 6.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 8.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 5.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.9 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 7.9 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 6.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.5 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 2.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.6 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 2.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 9.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |