Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxb7
Z-value: 3.83
Transcription factors associated with Hoxb7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb7
|
ENSRNOG00000007611 | homeo box B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb7 | rn6_v1_chr10_+_84135116_84135116 | 0.06 | 2.8e-01 | Click! |
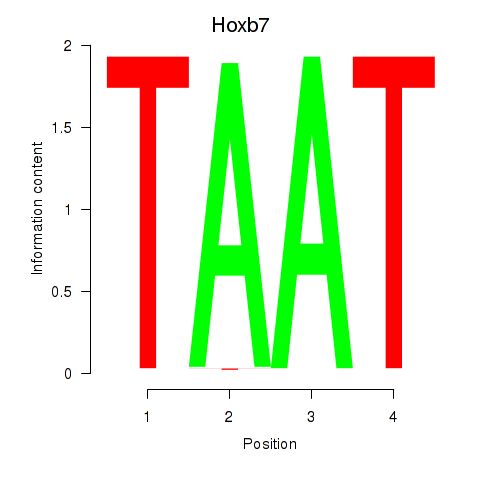
Activity profile of Hoxb7 motif
Sorted Z-values of Hoxb7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_46731403 | 269.20 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr16_-_39476384 | 234.71 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chrX_-_142164220 | 206.12 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr1_-_93949187 | 193.54 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr11_-_4397361 | 180.77 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr16_-_39476025 | 180.55 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr6_+_48452369 | 179.34 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr6_+_64789940 | 176.90 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr3_+_48096954 | 162.65 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr3_-_52447622 | 157.66 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr2_+_217963456 | 155.31 |
ENSRNOT00000024243
|
Olfm3
|
olfactomedin 3 |
| chr9_-_32019205 | 153.80 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr5_+_120340646 | 153.32 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr1_+_240355149 | 148.88 |
ENSRNOT00000018521
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr10_-_8654892 | 142.27 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr2_-_89310946 | 139.45 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr3_-_48831417 | 130.38 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr3_+_48082935 | 129.99 |
ENSRNOT00000087711
ENSRNOT00000067545 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr4_-_85915099 | 129.66 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr12_-_29743705 | 128.43 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr4_-_40385349 | 124.26 |
ENSRNOT00000039005
|
Gpr85
|
G protein-coupled receptor 85 |
| chr10_-_27862868 | 124.18 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr8_-_47094352 | 123.44 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chr17_+_63635086 | 123.06 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr9_-_30844199 | 121.56 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr10_-_27179900 | 120.78 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr15_-_77736892 | 120.66 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr3_-_26056818 | 120.36 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr3_-_44086006 | 119.85 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr14_-_43143973 | 118.07 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr7_-_50278842 | 117.52 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr3_-_66417741 | 113.19 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr3_+_48106099 | 111.89 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr20_-_25826658 | 106.13 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr2_+_144861455 | 105.67 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr4_+_172942020 | 102.37 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr18_-_37096132 | 101.58 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr4_-_55011415 | 99.58 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr15_+_11298478 | 98.22 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr1_-_279277339 | 97.91 |
ENSRNOT00000023667
|
Gfra1
|
GDNF family receptor alpha 1 |
| chr2_+_102685513 | 97.55 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr15_-_93307420 | 92.74 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr10_+_99388130 | 92.72 |
ENSRNOT00000006238
|
Kcnj16
|
potassium voltage-gated channel subfamily J member 16 |
| chr2_-_173087648 | 91.51 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr2_-_41784929 | 91.51 |
ENSRNOT00000086851
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr11_-_27971359 | 90.23 |
ENSRNOT00000085629
ENSRNOT00000051060 ENSRNOT00000042581 ENSRNOT00000050073 ENSRNOT00000081066 |
Grik1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr19_+_6046665 | 89.51 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr2_-_5577369 | 89.51 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_-_51612397 | 89.27 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr1_+_27476375 | 87.45 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr2_+_66940057 | 85.91 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr10_-_8498422 | 84.01 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr4_-_135069970 | 83.78 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr9_+_73378057 | 83.55 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr9_+_73334618 | 83.19 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr15_+_38341089 | 83.03 |
ENSRNOT00000015367
|
Fgf9
|
fibroblast growth factor 9 |
| chr1_-_173764246 | 82.69 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr2_-_63166509 | 82.22 |
ENSRNOT00000018246
|
Cdh6
|
cadherin 6 |
| chrX_+_33884499 | 81.80 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr15_+_104026601 | 81.27 |
ENSRNOT00000013520
ENSRNOT00000083269 ENSRNOT00000093403 |
Cldn10
|
claudin 10 |
| chr15_-_14737704 | 80.57 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr20_+_25990304 | 80.49 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr7_-_83670356 | 80.45 |
ENSRNOT00000005584
|
Sybu
|
syntabulin |
| chr3_+_47677720 | 80.28 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr2_-_35104963 | 78.88 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr7_-_92996025 | 78.20 |
ENSRNOT00000076327
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr2_+_145174876 | 77.99 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr15_+_96821832 | 76.94 |
ENSRNOT00000012785
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr17_+_47721977 | 76.71 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr5_-_109651730 | 76.57 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr14_+_66598259 | 76.19 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr1_+_106998623 | 75.79 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr12_-_35979193 | 74.81 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_+_69415057 | 73.78 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr4_+_94696965 | 73.78 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chrX_-_32153794 | 73.74 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr3_+_51687809 | 73.67 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr14_+_114152472 | 73.08 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr10_-_27366665 | 71.82 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr1_+_234363994 | 71.40 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr9_+_73418607 | 71.15 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr15_+_17775692 | 70.76 |
ENSRNOT00000061169
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr2_-_231648122 | 70.70 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr10_-_27179254 | 70.35 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr6_+_83421882 | 69.14 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr14_+_39964588 | 68.77 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr4_+_110699557 | 68.62 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chrX_+_6791136 | 68.04 |
ENSRNOT00000003984
|
LOC100909913
|
norrin-like |
| chrX_+_23081125 | 67.88 |
ENSRNOT00000071639
|
AABR07037520.1
|
|
| chrX_-_142248369 | 67.87 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr15_+_1054937 | 67.66 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr9_-_85243001 | 67.63 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr18_-_6782757 | 67.31 |
ENSRNOT00000068150
|
Aqp4
|
aquaporin 4 |
| chr2_-_184263564 | 66.62 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr6_-_8344574 | 66.61 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr2_+_72006099 | 66.48 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr9_+_86874685 | 66.38 |
ENSRNOT00000041337
|
NEWGENE_1305560
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr9_+_118849302 | 66.36 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr2_+_11658568 | 66.20 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chrX_-_29648359 | 66.19 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr5_+_29538380 | 65.97 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr15_+_17834635 | 65.84 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr4_+_22859622 | 65.64 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr5_-_12526962 | 64.95 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr7_-_76294663 | 64.89 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr7_+_133457255 | 64.80 |
ENSRNOT00000080621
|
Cntn1
|
contactin 1 |
| chr7_-_66172360 | 64.73 |
ENSRNOT00000005537
|
Fam19a2
|
family with sequence similarity 19 member A2, C-C motif chemokine like |
| chr5_-_168734296 | 64.10 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr20_-_54517709 | 63.98 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr13_+_106463368 | 63.62 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chr8_-_120446455 | 63.60 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr7_-_29701586 | 63.24 |
ENSRNOT00000009084
ENSRNOT00000089269 |
Ano4
|
anoctamin 4 |
| chr2_-_95472115 | 62.79 |
ENSRNOT00000015720
|
Stmn2
|
stathmin 2 |
| chr18_-_24182012 | 62.78 |
ENSRNOT00000023594
|
Syt4
|
synaptotagmin 4 |
| chr3_-_158328881 | 62.38 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr2_+_116970344 | 62.34 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr4_+_24222500 | 61.65 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr4_-_15859132 | 60.66 |
ENSRNOT00000082161
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chrX_+_118197217 | 60.32 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr14_-_28856214 | 59.85 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr1_+_268189277 | 59.56 |
ENSRNOT00000065001
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr6_+_8284878 | 59.46 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr4_+_136512201 | 59.39 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chr5_-_105579959 | 59.33 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr5_+_22392732 | 59.20 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr20_+_20378861 | 59.04 |
ENSRNOT00000091044
|
Ank3
|
ankyrin 3 |
| chr8_-_84320714 | 58.93 |
ENSRNOT00000079356
ENSRNOT00000088487 |
Tinag
|
tubulointerstitial nephritis antigen |
| chr3_-_25212049 | 58.07 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_+_198383201 | 57.44 |
ENSRNOT00000037405
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr4_+_138269142 | 57.11 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chr2_-_57935334 | 57.09 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr13_-_49487892 | 56.51 |
ENSRNOT00000044972
|
Nfasc
|
neurofascin |
| chr5_+_22380334 | 56.41 |
ENSRNOT00000009744
|
Clvs1
|
clavesin 1 |
| chrX_+_53053609 | 56.24 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr2_-_179704629 | 56.15 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr13_-_102857551 | 56.14 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr4_+_64088900 | 55.90 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr6_-_23291568 | 55.88 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr13_-_107886476 | 55.54 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr8_-_87419564 | 55.42 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr9_-_60672246 | 54.58 |
ENSRNOT00000017761
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr8_-_14880644 | 54.45 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr2_+_18354542 | 54.28 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr4_+_70252366 | 54.25 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr12_-_5682608 | 54.12 |
ENSRNOT00000076483
|
Fry
|
FRY microtubule binding protein |
| chr2_+_74360622 | 53.54 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr8_+_82038967 | 53.45 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chrM_+_7919 | 53.41 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chrM_+_10160 | 53.30 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_-_115175299 | 53.28 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr9_+_77834091 | 53.04 |
ENSRNOT00000033459
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr10_-_66848388 | 52.97 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr2_-_111793326 | 52.53 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chr18_+_62852303 | 52.27 |
ENSRNOT00000087673
|
Gnal
|
G protein subunit alpha L |
| chr17_+_41798783 | 52.14 |
ENSRNOT00000023519
|
Nrsn1
|
neurensin 1 |
| chr6_-_114476723 | 52.00 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr8_-_30222036 | 51.94 |
ENSRNOT00000035454
|
Ntm
|
neurotrimin |
| chr7_+_44009069 | 51.83 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr4_-_21668570 | 51.68 |
ENSRNOT00000007723
|
RGD1563349
|
similar to RIKEN cDNA 9330182L06 |
| chr12_-_10335499 | 51.52 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr1_+_59156251 | 50.74 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr11_+_20474483 | 50.72 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr15_+_56666012 | 50.69 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr10_-_21265026 | 50.63 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr1_-_43638161 | 50.05 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_+_256955652 | 49.74 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr13_-_95250235 | 49.69 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr14_-_6679878 | 49.59 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr17_+_11683862 | 49.36 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr4_+_21462779 | 49.16 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr3_-_46726946 | 49.12 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr18_-_14016713 | 48.90 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr2_-_184289126 | 48.73 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr3_-_57957346 | 48.72 |
ENSRNOT00000036728
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr18_+_65285318 | 48.64 |
ENSRNOT00000020431
|
Tcf4
|
transcription factor 4 |
| chrX_+_84064427 | 48.54 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr5_-_60655985 | 48.54 |
ENSRNOT00000093103
|
Fbxo10
|
F-box protein 10 |
| chr4_+_139670092 | 48.45 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr5_-_12563429 | 48.39 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chrM_+_7758 | 48.17 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr7_-_58149061 | 48.17 |
ENSRNOT00000005157
|
Tph2
|
tryptophan hydroxylase 2 |
| chr9_+_79630604 | 48.12 |
ENSRNOT00000021539
ENSRNOT00000083979 |
Tmem169
|
transmembrane protein 169 |
| chr13_-_76049363 | 48.01 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr9_-_19613360 | 47.61 |
ENSRNOT00000029593
|
Rcan2
|
regulator of calcineurin 2 |
| chr3_-_66885085 | 47.16 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr7_-_44121130 | 47.13 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr18_-_77579969 | 47.11 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr3_-_51643140 | 46.95 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr2_+_263212051 | 46.55 |
ENSRNOT00000089396
|
Negr1
|
neuronal growth regulator 1 |
| chr15_+_4209703 | 46.49 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr14_-_66978499 | 46.11 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr13_+_24823488 | 46.07 |
ENSRNOT00000019907
|
Cdh20
|
cadherin 20 |
| chr2_-_88553086 | 45.92 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr6_-_51019407 | 45.61 |
ENSRNOT00000011659
|
Gpr22
|
G protein-coupled receptor 22 |
| chr18_+_30581530 | 45.60 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr2_+_3662763 | 45.39 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr4_-_86466797 | 45.35 |
ENSRNOT00000017531
|
Pde1c
|
phosphodiesterase 1C |
| chr2_-_88763733 | 44.80 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr1_-_16203838 | 44.70 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr10_-_91661558 | 44.51 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 41.2 | 288.7 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 38.4 | 115.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 38.3 | 153.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 37.0 | 111.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 33.5 | 167.3 | GO:0061743 | motor learning(GO:0061743) |
| 30.1 | 331.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 29.2 | 87.6 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 28.3 | 113.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 26.5 | 105.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 26.3 | 52.5 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 25.9 | 233.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 24.1 | 457.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 22.6 | 113.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 22.0 | 154.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 21.1 | 63.4 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 21.1 | 63.2 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 20.8 | 83.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 20.4 | 183.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 19.6 | 19.6 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 19.2 | 153.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 18.9 | 189.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 18.8 | 262.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 18.7 | 56.2 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 18.5 | 55.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 18.5 | 129.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 17.5 | 105.3 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 17.5 | 52.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 16.8 | 67.3 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 16.8 | 33.6 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 16.5 | 66.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 16.5 | 49.6 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 16.5 | 165.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 16.5 | 66.0 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 16.5 | 66.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 16.5 | 49.4 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 16.1 | 48.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 15.8 | 157.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 15.2 | 60.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 15.2 | 75.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 14.8 | 74.0 | GO:0036233 | glycine import(GO:0036233) |
| 14.8 | 59.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 14.4 | 86.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 13.7 | 41.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 13.5 | 54.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 13.5 | 148.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 13.3 | 106.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 12.9 | 12.9 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 12.8 | 191.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 12.7 | 241.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 12.6 | 63.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 12.6 | 62.8 | GO:2000301 | negative regulation of vesicle fusion(GO:0031339) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 11.7 | 70.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 11.7 | 35.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 11.4 | 34.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 11.3 | 440.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 11.3 | 90.2 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 11.0 | 44.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 10.8 | 54.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 10.7 | 21.4 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 10.5 | 73.7 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 10.5 | 31.5 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 10.5 | 167.5 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 10.3 | 123.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 10.3 | 30.8 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 10.2 | 51.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 10.1 | 60.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 10.1 | 30.2 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 10.0 | 40.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 10.0 | 30.0 | GO:0015827 | angiotensin-mediated drinking behavior(GO:0003051) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 9.8 | 49.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 9.7 | 29.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 9.7 | 19.3 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 9.6 | 38.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 9.5 | 28.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 9.4 | 37.8 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 9.4 | 47.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 9.3 | 37.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 9.0 | 26.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 9.0 | 62.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 8.9 | 35.8 | GO:0031179 | peptide modification(GO:0031179) |
| 8.8 | 79.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 8.8 | 43.8 | GO:0060023 | soft palate development(GO:0060023) |
| 8.7 | 34.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 8.7 | 52.0 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 8.6 | 60.5 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 8.5 | 299.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 8.5 | 33.9 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 8.4 | 42.0 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 8.3 | 25.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 8.3 | 66.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 8.3 | 49.7 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 8.3 | 24.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 8.2 | 41.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 8.0 | 16.0 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 7.9 | 31.5 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 7.9 | 31.5 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 7.8 | 1098.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 7.8 | 62.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 7.7 | 23.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 7.6 | 38.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 7.6 | 128.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 7.5 | 246.7 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 7.4 | 22.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 7.3 | 101.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 7.2 | 21.7 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 7.2 | 129.7 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 7.1 | 149.6 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 7.1 | 92.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 7.1 | 28.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 6.9 | 75.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 6.8 | 20.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 6.7 | 13.5 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 6.7 | 26.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 6.6 | 39.8 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 6.5 | 71.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 6.5 | 19.4 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 6.2 | 43.7 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 6.2 | 18.7 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 6.2 | 31.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 6.1 | 42.9 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 6.0 | 162.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 6.0 | 18.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 6.0 | 23.9 | GO:0071651 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 6.0 | 17.9 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 5.9 | 41.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 5.9 | 342.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 5.7 | 131.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 5.7 | 11.5 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 5.7 | 22.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 5.6 | 61.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 5.6 | 78.0 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 5.5 | 32.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 5.4 | 32.4 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 5.2 | 15.7 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 5.2 | 26.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 5.2 | 15.5 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 5.2 | 10.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 5.1 | 30.8 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 5.1 | 10.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 5.1 | 20.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 5.0 | 169.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 4.9 | 14.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 4.8 | 43.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 4.8 | 154.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 4.7 | 18.9 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 4.7 | 23.5 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 4.6 | 9.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 4.5 | 72.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 4.5 | 9.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 4.5 | 63.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 4.4 | 123.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 4.4 | 122.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 4.4 | 26.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 4.4 | 26.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 4.4 | 30.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 4.4 | 21.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 4.3 | 4.3 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 4.3 | 13.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 4.3 | 17.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 4.3 | 12.8 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 4.3 | 59.6 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 4.2 | 38.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 4.2 | 21.1 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 4.2 | 16.7 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 4.1 | 8.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 4.0 | 8.0 | GO:2000078 | positive regulation of type B pancreatic cell development(GO:2000078) |
| 4.0 | 44.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 4.0 | 11.9 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) response to formaldehyde(GO:1904404) |
| 3.9 | 27.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 3.9 | 11.8 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 3.9 | 39.3 | GO:2001053 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 3.9 | 125.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 3.9 | 38.8 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 3.9 | 30.8 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 3.8 | 57.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 3.7 | 11.2 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 3.7 | 18.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 3.7 | 82.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 3.6 | 50.7 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 3.6 | 35.6 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 3.5 | 10.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 3.4 | 24.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 3.4 | 16.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 3.3 | 30.0 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 3.3 | 36.4 | GO:0009642 | response to light intensity(GO:0009642) |
| 3.2 | 70.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 3.1 | 65.6 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 3.1 | 59.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 3.1 | 9.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 3.1 | 24.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 2.9 | 11.6 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 2.9 | 14.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 2.9 | 109.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 2.9 | 8.6 | GO:1904746 | glomerular endothelium development(GO:0072011) negative regulation of apoptotic process involved in morphogenesis(GO:1902338) negative regulation of apoptotic process involved in development(GO:1904746) |
| 2.9 | 28.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 2.8 | 41.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 2.7 | 13.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 2.7 | 13.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 2.7 | 29.2 | GO:0007602 | phototransduction(GO:0007602) |
| 2.5 | 10.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 2.5 | 14.9 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 2.5 | 131.0 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 2.5 | 34.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 2.5 | 22.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 2.4 | 24.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 2.4 | 4.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 2.4 | 28.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 2.3 | 7.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 2.3 | 9.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 2.2 | 28.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 2.1 | 87.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 2.1 | 18.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 2.1 | 53.5 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 2.0 | 4.0 | GO:1902276 | amylase secretion(GO:0036394) pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 2.0 | 50.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 1.9 | 13.5 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 1.9 | 26.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 1.9 | 7.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 1.9 | 9.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 1.8 | 14.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 1.7 | 12.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 1.7 | 19.1 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 1.7 | 92.5 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 1.7 | 28.8 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 1.7 | 16.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 1.6 | 19.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.6 | 9.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.6 | 7.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.6 | 3.1 | GO:1901963 | regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 1.6 | 9.4 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 1.5 | 13.8 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 1.5 | 10.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 1.5 | 3.0 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.5 | 14.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.5 | 8.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 1.5 | 11.6 | GO:0048840 | otolith development(GO:0048840) |
| 1.5 | 4.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 1.4 | 10.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 1.4 | 38.5 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 1.4 | 9.9 | GO:0071000 | response to magnetism(GO:0071000) |
| 1.4 | 11.2 | GO:0097369 | sodium ion import(GO:0097369) |
| 1.3 | 6.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 1.3 | 10.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 1.3 | 17.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.3 | 18.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.3 | 5.1 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 1.3 | 48.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 1.3 | 16.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 1.3 | 8.8 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 1.2 | 4.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.2 | 117.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 1.2 | 12.8 | GO:0030828 | positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 1.2 | 48.6 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 1.1 | 14.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 1.1 | 2.3 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 1.1 | 10.0 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 1.0 | 22.9 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 1.0 | 3.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 1.0 | 4.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 1.0 | 22.6 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 1.0 | 13.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 1.0 | 26.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 1.0 | 9.1 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 1.0 | 8.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 1.0 | 11.0 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 1.0 | 4.9 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 1.0 | 6.8 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.9 | 29.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.9 | 35.3 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.9 | 6.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.9 | 12.2 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.9 | 19.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.9 | 24.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.9 | 6.9 | GO:1900040 | regulation of interleukin-2 secretion(GO:1900040) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.9 | 21.4 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.8 | 99.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.8 | 19.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.8 | 3.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.8 | 35.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.8 | 50.3 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.8 | 3.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.8 | 3.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.8 | 40.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.8 | 20.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.7 | 18.0 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.7 | 4.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.7 | 9.5 | GO:0043586 | tongue development(GO:0043586) |
| 0.7 | 10.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.7 | 23.3 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.7 | 4.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.7 | 59.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.7 | 2.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.6 | 83.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.6 | 9.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.6 | 9.3 | GO:0098926 | postsynaptic signal transduction(GO:0098926) |
| 0.6 | 4.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.5 | 10.9 | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) |
| 0.5 | 12.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.5 | 1.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.5 | 7.5 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.5 | 46.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.5 | 31.2 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.4 | 1.7 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.4 | 1.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 1.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.4 | 29.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.4 | 3.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.4 | 0.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.4 | 4.4 | GO:0042534 | tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.4 | 1.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.4 | 3.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 2.4 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.3 | 25.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.3 | 3.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 13.7 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.3 | 4.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.3 | 23.6 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.3 | 8.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 25.6 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.3 | 3.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 0.8 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 4.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 8.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 10.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 17.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.2 | 2.8 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.2 | 20.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.2 | 17.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 8.5 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.2 | 4.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 2.8 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 4.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.2 | 6.0 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 0.5 | GO:0033386 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.1 | 0.7 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 0.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 5.9 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.1 | 2.7 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 2.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 1.3 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.2 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 2.1 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 8.1 | GO:0001822 | kidney development(GO:0001822) |
| 0.0 | 1.3 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.6 | GO:0009855 | specification of symmetry(GO:0009799) determination of bilateral symmetry(GO:0009855) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 71.4 | 642.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 46.3 | 277.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 27.8 | 83.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 23.1 | 115.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 20.0 | 119.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 19.4 | 523.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 19.3 | 116.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 16.2 | 243.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 15.0 | 344.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 14.6 | 43.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 12.7 | 38.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 12.7 | 76.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 12.3 | 37.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 12.1 | 386.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 11.4 | 466.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 9.6 | 143.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 9.6 | 9.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 9.3 | 55.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 9.3 | 130.4 | GO:0031045 | dense core granule(GO:0031045) |
| 9.1 | 618.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 8.9 | 80.4 | GO:0097433 | dense body(GO:0097433) |
| 8.8 | 70.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 8.2 | 123.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 7.8 | 156.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 7.7 | 46.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 7.1 | 78.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 7.0 | 118.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 6.9 | 55.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 6.8 | 41.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 6.6 | 257.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 6.2 | 73.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 6.1 | 164.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 5.6 | 581.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 5.6 | 16.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 5.5 | 77.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 5.1 | 10.2 | GO:1990357 | terminal web(GO:1990357) |
| 4.8 | 19.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 4.7 | 14.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 4.7 | 42.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 4.4 | 114.9 | GO:0030673 | axolemma(GO:0030673) |
| 4.4 | 52.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 4.3 | 221.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 4.3 | 21.7 | GO:0005940 | septin ring(GO:0005940) |
| 4.3 | 12.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 4.3 | 115.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 4.3 | 42.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 4.2 | 12.5 | GO:0044308 | axonal spine(GO:0044308) |
| 3.7 | 55.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 3.7 | 14.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 3.6 | 40.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 3.5 | 35.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 3.5 | 41.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 3.4 | 27.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 3.4 | 73.8 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 3.1 | 21.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 2.9 | 47.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 2.9 | 20.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 2.9 | 498.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 2.8 | 117.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 2.7 | 73.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 2.7 | 24.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 2.7 | 18.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 2.7 | 34.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 2.6 | 13.0 | GO:0070695 | FHF complex(GO:0070695) |
| 2.6 | 167.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 2.4 | 28.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 2.3 | 38.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 2.2 | 44.4 | GO:0010369 | chromocenter(GO:0010369) |
| 2.2 | 31.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 2.2 | 154.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 2.1 | 17.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 2.0 | 8.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 2.0 | 22.4 | GO:0043196 | varicosity(GO:0043196) |
| 2.0 | 340.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 2.0 | 13.8 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.9 | 22.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.8 | 272.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 1.7 | 27.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 1.7 | 20.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 1.7 | 30.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 1.7 | 11.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.6 | 67.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 1.6 | 49.1 | GO:0008305 | integrin complex(GO:0008305) |
| 1.6 | 4.8 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 1.6 | 167.5 | GO:0098794 | postsynapse(GO:0098794) |
| 1.5 | 16.2 | GO:0002177 | manchette(GO:0002177) |
| 1.5 | 4.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.5 | 11.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.4 | 33.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 1.3 | 10.0 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.2 | 34.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.2 | 137.7 | GO:0030427 | site of polarized growth(GO:0030427) |
| 1.2 | 988.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 1.2 | 20.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 1.2 | 13.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.2 | 10.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.1 | 11.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.1 | 100.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 1.1 | 68.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 1.1 | 24.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 1.0 | 3.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 1.0 | 6.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.0 | 4.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.0 | 13.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.9 | 76.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.9 | 48.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.9 | 214.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.8 | 7.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.8 | 24.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.8 | 37.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.8 | 18.6 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.8 | 10.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.8 | 16.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.7 | 22.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.7 | 14.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.7 | 4.5 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 45.2 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.7 | 113.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.7 | 2.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 8.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.6 | 9.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.6 | 51.0 | GO:0005814 | centriole(GO:0005814) |
| 0.6 | 4.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 13.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.6 | 18.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.5 | 31.1 | GO:0030424 | axon(GO:0030424) |
| 0.5 | 12.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 8.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.5 | 3.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.5 | 12.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.5 | 57.2 | GO:0045202 | synapse(GO:0045202) |
| 0.5 | 7.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.4 | 14.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 5.9 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.4 | 14.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 16.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.3 | 7.9 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 35.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 17.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 1.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.9 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.2 | 21.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 9.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 7.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 13.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 8.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.2 | 8.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 37.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 5.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 8.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 7.1 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.1 | 4.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 270.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 57.8 | 462.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 47.7 | 333.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 39.2 | 196.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 37.7 | 339.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 37.0 | 111.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 36.1 | 180.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 29.5 | 118.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 29.4 | 88.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 25.9 | 103.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 24.9 | 99.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 23.6 | 70.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 23.1 | 92.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 23.1 | 115.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 22.5 | 67.5 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 20.2 | 60.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 17.6 | 52.9 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 17.5 | 52.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 17.0 | 170.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 16.5 | 66.0 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 16.4 | 49.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 15.9 | 63.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 14.6 | 410.0 | GO:0031402 | sodium ion binding(GO:0031402) |
| 14.4 | 404.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 14.3 | 71.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 13.9 | 55.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 13.4 | 160.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 13.1 | 301.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 12.7 | 38.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 12.3 | 147.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 11.8 | 270.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 11.6 | 46.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 11.4 | 57.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 11.3 | 33.9 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 10.9 | 109.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 10.1 | 30.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 9.5 | 38.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 9.4 | 65.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 8.9 | 35.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 8.9 | 26.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 8.7 | 234.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 8.6 | 103.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 8.3 | 33.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 8.2 | 41.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 7.9 | 31.5 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 7.8 | 38.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 7.7 | 30.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 7.6 | 105.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 7.4 | 22.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 7.3 | 29.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 7.0 | 42.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 6.9 | 54.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 6.7 | 47.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 6.7 | 313.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 6.6 | 39.7 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 6.5 | 19.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 6.4 | 19.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 6.3 | 18.9 | GO:0031208 | POZ domain binding(GO:0031208) |
| 6.2 | 30.8 | GO:1902282 | A-type (transient outward) potassium channel activity(GO:0005250) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 6.1 | 72.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 5.7 | 40.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 5.6 | 163.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 5.6 | 22.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 5.5 | 210.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 5.5 | 49.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 5.5 | 125.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 5.2 | 62.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 5.2 | 41.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 5.2 | 15.5 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 5.1 | 20.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 5.1 | 66.4 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 4.9 | 73.8 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 4.9 | 233.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 4.8 | 115.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 4.8 | 14.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 4.8 | 67.3 | GO:0015250 | water channel activity(GO:0015250) |
| 4.8 | 28.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 4.7 | 28.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 4.7 | 56.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 4.6 | 68.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 4.5 | 103.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 4.4 | 31.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 4.4 | 30.5 | GO:0071253 | connexin binding(GO:0071253) |
| 4.3 | 12.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 4.3 | 439.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 4.0 | 11.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 3.9 | 31.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 3.9 | 31.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 3.7 | 63.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 3.7 | 37.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 3.5 | 10.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 3.4 | 17.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 3.4 | 10.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 3.4 | 24.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 3.4 | 27.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 3.3 | 16.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 3.3 | 46.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 3.3 | 19.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 3.2 | 19.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 3.2 | 34.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 3.1 | 9.3 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 3.1 | 70.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 3.0 | 9.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 3.0 | 174.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 3.0 | 81.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 2.8 | 39.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 2.7 | 29.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 2.7 | 8.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 2.5 | 48.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 2.5 | 10.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 2.5 | 9.9 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 2.5 | 17.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 2.5 | 9.9 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 2.4 | 22.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 2.4 | 70.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 2.4 | 19.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 2.3 | 7.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 2.3 | 99.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 2.3 | 46.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 2.2 | 62.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 2.2 | 211.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 2.2 | 15.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.2 | 39.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 2.2 | 54.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 2.1 | 29.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 2.1 | 6.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.0 | 10.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 2.0 | 18.0 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 2.0 | 35.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 2.0 | 56.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 1.9 | 28.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 1.9 | 41.4 | GO:0005112 | Notch binding(GO:0005112) |
| 1.9 | 167.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 1.8 | 25.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.8 | 9.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.8 | 28.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 1.8 | 23.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.8 | 24.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 1.7 | 10.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 1.7 | 86.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 1.7 | 25.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.6 | 96.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 1.6 | 7.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 1.5 | 35.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 1.5 | 16.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 1.5 | 7.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.5 | 11.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.4 | 18.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 1.4 | 30.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 1.4 | 11.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 1.4 | 52.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 1.4 | 11.0 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 1.3 | 18.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.3 | 118.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 1.3 | 32.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.3 | 67.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.3 | 812.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 1.3 | 6.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.3 | 27.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 1.2 | 15.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.2 | 4.8 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.2 | 25.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 1.2 | 39.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 1.1 | 17.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 1.0 | 13.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.0 | 277.8 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 1.0 | 8.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.0 | 23.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 1.0 | 14.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 1.0 | 4.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 6.7 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.9 | 45.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.9 | 9.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.9 | 4.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.9 | 29.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.9 | 48.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.9 | 21.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.8 | 89.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.8 | 7.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.8 | 4.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.8 | 11.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.8 | 6.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.8 | 20.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.7 | 22.8 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.7 | 5.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.7 | 65.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.7 | 511.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.7 | 11.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.7 | 6.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.6 | 4.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.6 | 21.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.6 | 9.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.6 | 5.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.6 | 10.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.6 | 4.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.5 | 77.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.5 | 92.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.5 | 13.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 1.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 4.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 29.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.4 | 15.1 | GO:0032934 | sterol binding(GO:0032934) |
| 0.4 | 39.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.4 | 3.5 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.4 | 1.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 3.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 12.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.4 | 1.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 7.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.3 | 6.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 6.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 7.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 12.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.3 | 18.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.3 | 12.7 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.3 | 18.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.3 | 6.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 8.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 63.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.2 | 27.7 | GO:0016874 | ligase activity(GO:0016874) |
| 0.2 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 1.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 42.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.2 | 10.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 28.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 1.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 19.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 99.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 3.8 | 97.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 3.4 | 133.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 3.1 | 90.2 | PID MYC PATHWAY | C-MYC pathway |
| 3.1 | 210.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 3.1 | 49.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 3.0 | 42.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 3.0 | 145.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 3.0 | 41.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 2.9 | 113.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 2.7 | 192.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 2.4 | 40.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 2.4 | 88.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 2.3 | 65.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 2.2 | 67.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 2.1 | 92.9 | PID ATR PATHWAY | ATR signaling pathway |
| 2.1 | 74.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 2.0 | 50.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 2.0 | 66.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 1.9 | 83.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 1.9 | 53.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 1.9 | 37.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 1.9 | 41.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 1.7 | 50.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.7 | 11.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 1.6 | 98.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 1.5 | 15.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 1.4 | 42.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 1.4 | 22.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 1.3 | 82.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 1.2 | 37.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 1.2 | 29.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 1.1 | 20.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.8 | 13.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.8 | 234.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.7 | 5.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.7 | 23.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.7 | 28.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.7 | 104.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.7 | 9.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.6 | 24.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.5 | 24.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 8.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 10.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 9.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 11.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 6.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 7.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.8 | 638.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 22.8 | 387.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 21.4 | 342.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 10.1 | 140.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 9.6 | 336.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 8.4 | 117.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 7.8 | 47.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 7.7 | 207.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 7.4 | 51.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 6.8 | 148.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 6.7 | 153.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 6.4 | 83.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 5.6 | 67.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 5.0 | 80.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 5.0 | 124.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 4.5 | 134.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 4.5 | 13.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 4.3 | 55.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 4.1 | 49.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 3.9 | 97.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 3.9 | 66.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 3.8 | 107.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 3.8 | 191.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 3.8 | 83.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 3.8 | 15.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 3.8 | 184.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 3.6 | 115.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 3.5 | 74.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 3.5 | 76.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 3.4 | 48.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 3.3 | 192.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 3.1 | 56.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 3.0 | 38.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 2.9 | 72.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 2.8 | 158.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 2.7 | 161.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 2.6 | 247.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 2.6 | 47.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 2.4 | 31.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 2.3 | 158.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 2.2 | 82.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 2.1 | 66.8 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 1.9 | 18.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.8 | 47.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 1.7 | 29.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.7 | 63.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 1.6 | 26.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 1.6 | 27.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.5 | 39.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 1.4 | 70.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 1.3 | 41.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 1.2 | 31.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 1.2 | 21.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.2 | 40.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.9 | 16.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.9 | 13.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.9 | 22.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.8 | 10.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.8 | 10.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.8 | 7.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.8 | 46.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.7 | 11.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.7 | 15.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.7 | 44.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.6 | 16.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.6 | 6.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 31.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.5 | 16.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.4 | 4.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 15.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.4 | 3.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 7.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 7.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 9.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.3 | 4.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 54.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 11.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 5.0 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.2 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 14.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 6.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |