Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
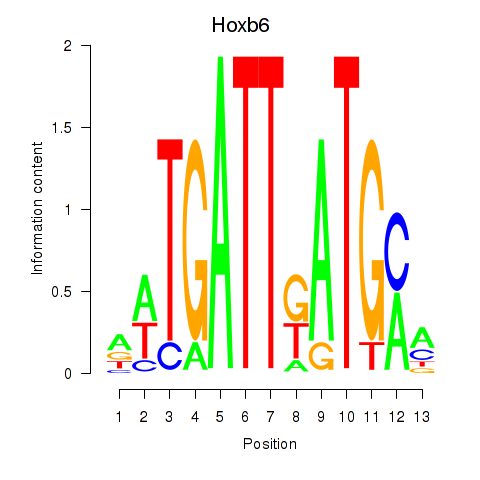
Results for Hoxb6
Z-value: 0.46
Transcription factors associated with Hoxb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb6
|
ENSRNOG00000007823 | homeo box B6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb6 | rn6_v1_chr10_+_84147836_84147836 | -0.41 | 1.2e-14 | Click! |
Activity profile of Hoxb6 motif
Sorted Z-values of Hoxb6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_85974644 | 13.52 |
ENSRNOT00000006098
ENSRNOT00000082974 |
Cacnb1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr17_-_43614844 | 11.91 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr6_-_95934296 | 9.54 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr9_-_63291350 | 7.05 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr11_-_45124423 | 6.71 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr18_-_786674 | 6.68 |
ENSRNOT00000021955
|
Cetn1
|
centrin 1 |
| chr1_-_197821936 | 6.24 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr14_+_39368530 | 5.95 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr3_+_110855000 | 5.68 |
ENSRNOT00000081613
|
Knl1
|
kinetochore scaffold 1 |
| chr13_+_78886163 | 4.83 |
ENSRNOT00000059266
ENSRNOT00000076513 |
Cenpl
|
centromere protein L |
| chr1_-_222182721 | 4.71 |
ENSRNOT00000078008
|
Tex40
|
testis expressed 40 |
| chr14_+_91557601 | 4.69 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr6_+_96007805 | 4.30 |
ENSRNOT00000010163
|
Mnat1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr13_-_91776397 | 4.27 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr9_-_42839837 | 4.22 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr13_-_61591139 | 4.05 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr5_-_156689415 | 3.91 |
ENSRNOT00000083474
|
Pink1
|
PTEN induced putative kinase 1 |
| chr5_+_48303366 | 3.69 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr10_-_56403188 | 3.41 |
ENSRNOT00000019947
|
Chrnb1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr7_+_35773928 | 3.17 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr11_+_53140599 | 2.94 |
ENSRNOT00000083438
|
Bbx
|
BBX, HMG-box containing |
| chr2_-_203350736 | 2.93 |
ENSRNOT00000090115
ENSRNOT00000084516 |
Ttf2
|
transcription termination factor 2 |
| chr19_-_37427989 | 2.91 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr4_-_177039677 | 2.57 |
ENSRNOT00000055538
ENSRNOT00000075667 |
RGD1561551
|
similar to Hypothetical protein MGC75664 |
| chr9_+_45605552 | 2.56 |
ENSRNOT00000059587
|
NMS
|
neuromedin S |
| chr4_-_28437676 | 2.49 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr5_+_8957405 | 2.45 |
ENSRNOT00000088164
|
Tcf24
|
transcription factor 24 |
| chr7_+_120923274 | 2.40 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr1_-_246010594 | 2.32 |
ENSRNOT00000023762
|
Rfx3
|
regulatory factor X3 |
| chr15_-_28935269 | 2.20 |
ENSRNOT00000060427
|
Olr1645
|
olfactory receptor 1645 |
| chrX_-_111102464 | 2.10 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr15_+_28439317 | 2.06 |
ENSRNOT00000060519
|
LOC690384
|
similar to ribosomal protein L31 |
| chr1_-_1889236 | 2.06 |
ENSRNOT00000087944
|
AABR07000159.2
|
|
| chr14_+_3882398 | 2.05 |
ENSRNOT00000033794
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr3_+_110508403 | 2.02 |
ENSRNOT00000079959
|
AABR07053500.2
|
|
| chr7_+_125288081 | 1.94 |
ENSRNOT00000085216
|
Parvg
|
parvin, gamma |
| chr9_+_53440272 | 1.79 |
ENSRNOT00000059159
|
LOC685203
|
hypothetical protein LOC685203 |
| chr17_+_69960160 | 1.70 |
ENSRNOT00000023887
|
Ucn3
|
urocortin 3 |
| chr4_-_164691405 | 1.64 |
ENSRNOT00000090979
ENSRNOT00000091932 ENSRNOT00000078219 |
Ly49s4
Ly49i2
|
Ly49 stimulatory receptor 4 Ly49 inhibitory receptor 2 |
| chr1_+_178039063 | 1.12 |
ENSRNOT00000046313
|
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr4_+_1841955 | 1.11 |
ENSRNOT00000019271
|
Olr1096
|
olfactory receptor 1096 |
| chr5_+_169898302 | 0.95 |
ENSRNOT00000087911
|
AABR07050561.1
|
|
| chr4_-_82133605 | 0.93 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr3_-_103128243 | 0.86 |
ENSRNOT00000046310
|
Olr779
|
olfactory receptor 779 |
| chrX_-_45284341 | 0.84 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr2_-_194875962 | 0.81 |
ENSRNOT00000085431
|
RGD1566337
|
similar to TDPOZ2 |
| chr17_-_27451832 | 0.76 |
ENSRNOT00000084417
|
Riok1
|
RIO kinase 1 |
| chr3_-_103400272 | 0.71 |
ENSRNOT00000046527
|
Olr790
|
olfactory receptor 790 |
| chr3_+_102723402 | 0.67 |
ENSRNOT00000040298
|
Olr765
|
olfactory receptor 765 |
| chr9_-_88748866 | 0.60 |
ENSRNOT00000071906
|
Rpl30l1
|
ribosomal protein L30-like 1 |
| chr3_+_103657115 | 0.56 |
ENSRNOT00000075514
|
Olr796
|
olfactory receptor 796 |
| chrX_-_83864150 | 0.56 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr9_-_52238564 | 0.55 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr3_-_103203299 | 0.53 |
ENSRNOT00000047208
|
Olr783
|
olfactory receptor 783 |
| chr8_+_42296303 | 0.52 |
ENSRNOT00000044272
|
Olr1238
|
olfactory receptor 1238 |
| chr1_+_79474809 | 0.50 |
ENSRNOT00000044927
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr5_+_160825419 | 0.46 |
ENSRNOT00000080134
|
AABR07050317.1
|
|
| chr3_-_103345333 | 0.44 |
ENSRNOT00000071479
|
LOC100909692
|
olfactory receptor 4F3/4F16/4F29-like |
| chr1_+_65292386 | 0.39 |
ENSRNOT00000029893
|
AABR07002044.1
|
|
| chr8_+_41911004 | 0.34 |
ENSRNOT00000071217
|
LOC100912067
|
olfactory receptor 143-like |
| chr1_-_172616633 | 0.32 |
ENSRNOT00000012919
|
Olr262
|
olfactory receptor 262 |
| chr3_-_75576520 | 0.26 |
ENSRNOT00000083330
|
Olr567
|
olfactory receptor 567 |
| chr11_+_43465370 | 0.23 |
ENSRNOT00000039939
|
Olr1547
|
olfactory receptor 1547 |
| chr13_-_98023829 | 0.22 |
ENSRNOT00000075426
|
Kif28p
|
kinesin family member 28, pseudogene |
| chr1_+_229480184 | 0.20 |
ENSRNOT00000044115
|
Olr387
|
olfactory receptor 387 |
| chr2_-_236149971 | 0.19 |
ENSRNOT00000089472
|
AABR07013412.1
|
|
| chr3_+_75999481 | 0.17 |
ENSRNOT00000074642
|
Olr595
|
olfactory receptor 595 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:2000729 | response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.8 | 3.9 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.8 | 2.3 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.4 | 6.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.4 | 3.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.4 | 5.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 4.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.3 | 13.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 3.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 1.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.2 | 11.9 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.1 | 2.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 2.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 6.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 3.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 2.4 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 | 0.8 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.7 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 5.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 4.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 2.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 2.9 | GO:0060348 | bone development(GO:0060348) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.4 | 11.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 5.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 13.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 3.8 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.2 | 3.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 2.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 3.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 6.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 5.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 9.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 2.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.1 | 4.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.7 | 13.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 1.7 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.4 | 3.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 9.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 3.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 3.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 5.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 11.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 6.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 2.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 4.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 14.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 3.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 6.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 5.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |