Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxb5
Z-value: 1.07
Transcription factors associated with Hoxb5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb5
|
ENSRNOG00000008010 | homeo box B5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb5 | rn6_v1_chr10_+_84152152_84152152 | -0.09 | 1.0e-01 | Click! |
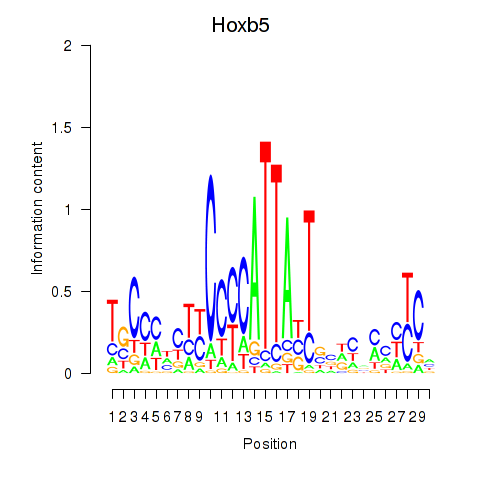
Activity profile of Hoxb5 motif
Sorted Z-values of Hoxb5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_164549455 | 169.93 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr15_-_33656089 | 64.53 |
ENSRNOT00000024186
|
Myh7
|
myosin heavy chain 7 |
| chr1_-_213650247 | 45.50 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr10_-_45480999 | 40.07 |
ENSRNOT00000078353
ENSRNOT00000084697 ENSRNOT00000087926 |
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr10_-_83898527 | 32.90 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr4_+_180291389 | 28.75 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr14_-_106393670 | 24.97 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chr1_+_126523169 | 21.78 |
ENSRNOT00000051000
|
Tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr13_-_82006005 | 19.74 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chrM_+_11736 | 19.57 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr20_+_37876650 | 18.64 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr11_+_58624198 | 17.95 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr1_+_94718402 | 17.18 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr16_+_21275311 | 15.72 |
ENSRNOT00000027980
|
LOC100911483
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-like |
| chr6_+_64789940 | 15.62 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr2_-_200762492 | 14.70 |
ENSRNOT00000056172
|
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr7_-_143420027 | 14.44 |
ENSRNOT00000082863
|
Krt2
|
keratin 2 |
| chrX_+_105537602 | 14.15 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr2_-_200762206 | 14.07 |
ENSRNOT00000068511
ENSRNOT00000086835 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr15_-_110046687 | 13.77 |
ENSRNOT00000057404
ENSRNOT00000006624 ENSRNOT00000089695 |
Nalcn
|
sodium leak channel, non-selective |
| chrX_-_14220662 | 13.61 |
ENSRNOT00000045753
|
Srpx
|
sushi-repeat-containing protein, X-linked |
| chr1_+_101012822 | 13.51 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chrM_+_5323 | 13.38 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr10_-_13115294 | 12.47 |
ENSRNOT00000005899
|
NEWGENE_6497122
|
FLYWCH family member 2 |
| chr4_+_175431904 | 10.99 |
ENSRNOT00000032843
|
Pde3a
|
phosphodiesterase 3A |
| chr4_+_88584242 | 10.92 |
ENSRNOT00000008973
|
Pyurf
|
PIGY upstream reading frame |
| chr3_-_15376767 | 10.38 |
ENSRNOT00000044885
|
Ndufa8
|
NADH:ubiquinone oxidoreductase subunit A8 |
| chr5_+_104362971 | 10.21 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr14_-_6679878 | 10.15 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr13_-_109997092 | 9.35 |
ENSRNOT00000005190
|
Nenf
|
neudesin neurotrophic factor |
| chr16_-_69961162 | 8.89 |
ENSRNOT00000074955
|
Prrg3
|
proline rich and Gla domain 3 |
| chr1_+_227892956 | 8.84 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr18_+_25163561 | 8.75 |
ENSRNOT00000042287
ENSRNOT00000049898 ENSRNOT00000017573 |
Bin1
|
bridging integrator 1 |
| chr4_+_49369296 | 8.70 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr5_-_28349927 | 8.60 |
ENSRNOT00000008725
|
Otud6b
|
OTU domain containing 6B |
| chr5_+_28395296 | 7.93 |
ENSRNOT00000009375
|
Tmem55a
|
transmembrane protein 55A |
| chrX_-_1718637 | 7.89 |
ENSRNOT00000064309
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr1_-_164307084 | 7.82 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr6_+_12415805 | 7.41 |
ENSRNOT00000022380
|
Gtf2a1l
|
general transcription factor 2A subunit 1 like |
| chr1_-_173682226 | 7.38 |
ENSRNOT00000020137
|
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr1_+_84304228 | 7.24 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr14_-_60964324 | 6.65 |
ENSRNOT00000005155
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr2_-_32518643 | 6.64 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr15_+_42653148 | 6.53 |
ENSRNOT00000022095
|
Clu
|
clusterin |
| chr18_+_30885789 | 6.45 |
ENSRNOT00000044434
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr12_-_38782010 | 6.37 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr12_+_2170630 | 6.24 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chr20_+_12052693 | 6.02 |
ENSRNOT00000083851
|
Adarb1
|
adenosine deaminase, RNA-specific, B1 |
| chr2_-_27287605 | 5.97 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chrX_-_106576314 | 5.26 |
ENSRNOT00000037823
|
Nxf3
|
nuclear RNA export factor 3 |
| chrX_+_106306795 | 5.06 |
ENSRNOT00000073661
|
Gprasp1
|
G protein-coupled receptor associated sorting protein 1 |
| chr6_-_139973811 | 5.05 |
ENSRNOT00000082875
|
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr9_+_16612433 | 4.90 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr5_+_144779681 | 4.85 |
ENSRNOT00000016493
|
LOC100294508
|
dyslexia susceptibility 2-like |
| chrX_+_151103576 | 4.80 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr1_-_168644038 | 4.79 |
ENSRNOT00000021304
|
Olr109
|
olfactory receptor 109 |
| chr15_-_12889102 | 4.61 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chr3_-_77402579 | 4.45 |
ENSRNOT00000043801
|
Olr658
|
olfactory receptor 658 |
| chr5_-_144779212 | 4.20 |
ENSRNOT00000016230
|
Ncdn
|
neurochondrin |
| chr3_+_92640752 | 4.13 |
ENSRNOT00000007604
|
Slc1a2
|
solute carrier family 1 member 2 |
| chr1_-_48825364 | 4.08 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr7_-_24102761 | 4.03 |
ENSRNOT00000008596
|
Btbd11
|
BTB domain containing 11 |
| chr3_+_73352352 | 3.82 |
ENSRNOT00000012814
|
Olr472
|
olfactory receptor 472 |
| chr1_-_107385257 | 3.80 |
ENSRNOT00000074296
|
Ccdc179
|
coiled-coil domain containing 179 |
| chr2_+_187740531 | 3.41 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr12_-_52124779 | 3.28 |
ENSRNOT00000088839
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chrX_-_100632562 | 3.23 |
ENSRNOT00000073416
|
LOC102555578
|
zinc finger protein 120-like |
| chr3_+_145882819 | 3.07 |
ENSRNOT00000042110
|
LOC102549291
|
zinc finger protein 120-like |
| chr2_+_196608496 | 3.06 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr6_+_27536184 | 3.03 |
ENSRNOT00000032829
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr10_+_34631398 | 2.80 |
ENSRNOT00000046027
|
Olr1391
|
olfactory receptor 1391 |
| chr5_-_38923095 | 2.77 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chrX_-_61151601 | 2.75 |
ENSRNOT00000004132
|
Mageb4
|
melanoma antigen family B, 4 |
| chr3_-_176465162 | 2.59 |
ENSRNOT00000048807
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr1_+_172813394 | 2.55 |
ENSRNOT00000090059
|
Olr272
|
olfactory receptor 272 |
| chr8_+_42046393 | 2.31 |
ENSRNOT00000072353
|
Olr1229
|
olfactory receptor 1229 |
| chr6_+_135866739 | 2.26 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr17_+_4270207 | 2.26 |
ENSRNOT00000061391
|
RGD1308751
|
similar to Cathepsin L precursor (Major excreted protein) (MEP) |
| chrX_+_105427939 | 2.20 |
ENSRNOT00000057217
|
LOC501618
|
LRRGT00179 |
| chr17_+_4256299 | 2.16 |
ENSRNOT00000083451
ENSRNOT00000061398 ENSRNOT00000090013 |
RGD1308751
|
similar to Cathepsin L precursor (Major excreted protein) (MEP) |
| chr10_+_48903540 | 2.04 |
ENSRNOT00000004248
|
Trpv2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr20_+_5067330 | 1.97 |
ENSRNOT00000037482
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr6_+_112203679 | 1.95 |
ENSRNOT00000031205
|
Nrxn3
|
neurexin 3 |
| chr8_+_119261747 | 1.83 |
ENSRNOT00000079648
|
AC132539.1
|
|
| chr12_+_41463922 | 1.74 |
ENSRNOT00000089378
|
Cfap73
|
cilia and flagella associated protein 73 |
| chr1_+_72545596 | 1.71 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr10_-_87891285 | 1.70 |
ENSRNOT00000090052
|
Rn50_10_0877.5
|
|
| chr7_+_29282305 | 1.65 |
ENSRNOT00000007623
|
Arl1
|
ADP-ribosylation factor like GTPase 1 |
| chr1_+_169240508 | 1.65 |
ENSRNOT00000022759
|
Olr143
|
olfactory receptor 143 |
| chr15_-_36321572 | 1.53 |
ENSRNOT00000073934
|
AABR07018016.1
|
|
| chr3_-_76708430 | 1.48 |
ENSRNOT00000007890
|
Olr630
|
olfactory receptor 630 |
| chr2_-_242828629 | 1.44 |
ENSRNOT00000043369
|
LOC499715
|
LRRGT00095 |
| chr19_+_54493483 | 1.43 |
ENSRNOT00000078002
|
AABR07044063.1
|
|
| chr1_-_65058502 | 1.36 |
ENSRNOT00000092003
|
AABR07002025.1
|
|
| chr9_+_12633990 | 1.33 |
ENSRNOT00000066517
ENSRNOT00000077532 |
Dazl
|
deleted in azoospermia-like |
| chr1_-_150210403 | 1.30 |
ENSRNOT00000081156
|
Olr29
|
olfactory receptor 29 |
| chr1_-_168029293 | 1.25 |
ENSRNOT00000020699
|
Olr49
|
olfactory receptor 49 |
| chr1_-_82580007 | 1.24 |
ENSRNOT00000078668
|
Axl
|
Axl receptor tyrosine kinase |
| chr16_-_2048292 | 1.20 |
ENSRNOT00000061883
|
Zcchc24
|
zinc finger CCHC-type containing 24 |
| chr1_+_89202527 | 1.19 |
ENSRNOT00000028526
|
Sbsn
|
suprabasin |
| chr10_+_103349172 | 1.15 |
ENSRNOT00000035963
|
Gpr142
|
G protein-coupled receptor 142 |
| chr11_+_70056624 | 1.13 |
ENSRNOT00000002447
|
AC133403.1
|
|
| chr9_+_9842585 | 1.13 |
ENSRNOT00000070806
|
Cd70
|
Cd70 molecule |
| chr20_+_10123651 | 1.11 |
ENSRNOT00000001559
|
Pde9a
|
phosphodiesterase 9A |
| chr1_+_150225373 | 1.10 |
ENSRNOT00000051266
|
Olr30
|
olfactory receptor 30 |
| chr1_+_169741497 | 1.06 |
ENSRNOT00000072865
|
Olr164
|
olfactory receptor 164 |
| chr20_+_4162055 | 1.05 |
ENSRNOT00000081236
|
Btnl3
|
butyrophilin-like 3 |
| chr1_+_60248695 | 1.02 |
ENSRNOT00000079558
|
Vom1r9
|
vomeronasal 1 receptor 9 |
| chr14_-_86868598 | 0.98 |
ENSRNOT00000087212
|
Nacad
|
NAC alpha domain containing |
| chr3_-_4405194 | 0.92 |
ENSRNOT00000072025
|
AABR07051266.1
|
|
| chr4_+_1400301 | 0.90 |
ENSRNOT00000074022
|
LOC100911888
|
olfactory receptor 143-like |
| chr17_+_57906050 | 0.83 |
ENSRNOT00000077338
|
AABR07028157.1
|
|
| chr3_+_16039249 | 0.59 |
ENSRNOT00000046196
|
Olr403
|
olfactory receptor 403 |
| chr2_-_188471988 | 0.54 |
ENSRNOT00000027785
|
Hcn3
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 3 |
| chr1_-_150173316 | 0.50 |
ENSRNOT00000044676
|
Olr27
|
olfactory receptor 27 |
| chr1_-_82279145 | 0.37 |
ENSRNOT00000057433
|
Cxcl17
|
C-X-C motif chemokine ligand 17 |
| chr14_-_35652709 | 0.20 |
ENSRNOT00000003080
|
Gsx2
|
GS homeobox 2 |
| chrX_-_30831483 | 0.18 |
ENSRNOT00000004443
|
Glra2
|
glycine receptor, alpha 2 |
| chrX_+_124894466 | 0.02 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr19_-_52186260 | 0.00 |
ENSRNOT00000020838
|
Mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.5 | 64.5 | GO:1905243 | slow-twitch skeletal muscle fiber contraction(GO:0031444) response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) |
| 17.0 | 169.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 7.3 | 21.8 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) threonyl-tRNA aminoacylation(GO:0006435) |
| 6.6 | 19.7 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 6.2 | 18.6 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 4.9 | 58.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 4.8 | 28.8 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 3.4 | 10.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 3.1 | 15.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 2.9 | 8.7 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 2.4 | 7.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 2.3 | 13.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 2.2 | 11.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 2.2 | 6.5 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.1 | 17.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 2.1 | 25.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 2.0 | 6.0 | GO:0061744 | motor behavior(GO:0061744) |
| 2.0 | 13.8 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 1.8 | 17.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.7 | 15.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.2 | 8.7 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.2 | 32.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.8 | 14.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.8 | 6.7 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.8 | 4.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 9.4 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.5 | 6.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.5 | 4.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 19.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.4 | 5.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 1.2 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.3 | 13.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.3 | 7.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.3 | 40.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.2 | 6.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 5.1 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 1.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 3.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.2 | 6.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 7.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 2.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.0 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 1.3 | GO:0007135 | meiosis II(GO:0007135) |
| 0.1 | 7.9 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 2.0 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 7.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.5 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 4.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 4.2 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 4.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 2.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 2.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 24.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) cellular response to zinc ion(GO:0071294) |
| 0.0 | 3.3 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 64.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 3.7 | 58.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 2.5 | 32.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 2.5 | 20.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.7 | 21.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.7 | 40.1 | GO:0031430 | M band(GO:0031430) |
| 1.6 | 17.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 1.5 | 28.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.2 | 7.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.2 | 10.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.0 | 18.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.9 | 167.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.9 | 45.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.9 | 24.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.7 | 6.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.6 | 14.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 6.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 4.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 32.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 10.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 7.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 7.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.2 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 7.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 8.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 6.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 7.1 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 12.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.0 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 4.9 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.8 | GO:0004829 | proline-tRNA ligase activity(GO:0004827) threonine-tRNA ligase activity(GO:0004829) |
| 6.6 | 19.7 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 6.2 | 25.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 5.8 | 28.8 | GO:0030283 | testosterone dehydrogenase [NAD(P)] activity(GO:0030283) |
| 4.6 | 64.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 4.5 | 17.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 3.7 | 18.6 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 2.9 | 14.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 2.9 | 40.1 | GO:0031432 | titin binding(GO:0031432) |
| 2.1 | 17.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 2.0 | 58.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.7 | 15.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.7 | 13.8 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 1.6 | 32.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.6 | 7.9 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 1.5 | 6.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 1.3 | 6.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.3 | 35.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.0 | 3.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.8 | 4.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.6 | 11.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.5 | 9.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.4 | 6.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 8.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 4.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 8.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 13.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 14.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 10.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 132.7 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.1 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.2 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.1 | 9.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 4.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.1 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 3.4 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 7.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 10.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 4.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 13.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 6.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 36.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.6 | 40.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.4 | 10.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 13.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 8.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 6.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 16.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 13.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 4.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 28.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 2.1 | 18.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 2.1 | 32.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.7 | 13.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.7 | 40.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.6 | 25.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 12.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 15.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 27.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 4.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 10.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 4.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 8.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 3.6 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |