Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxb4
Z-value: 0.73
Transcription factors associated with Hoxb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb4
|
ENSRNOG00000008191 | homeo box B4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb4 | rn6_v1_chr10_+_84167331_84167331 | 0.03 | 6.3e-01 | Click! |
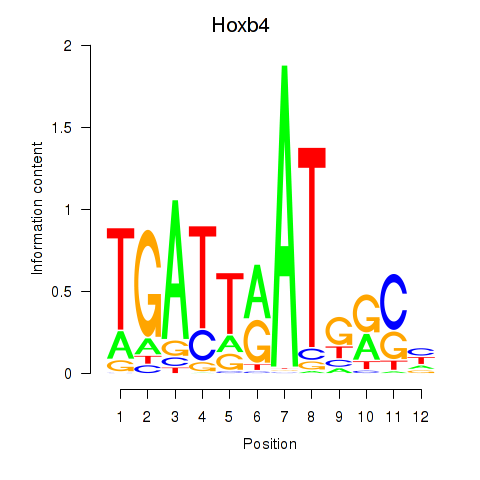
Activity profile of Hoxb4 motif
Sorted Z-values of Hoxb4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_26999795 | 45.95 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr7_-_29171783 | 36.23 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr3_-_23020441 | 25.70 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr11_+_17538063 | 24.11 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr19_-_37427989 | 23.04 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_-_215838209 | 22.28 |
ENSRNOT00000050760
|
Igf2
|
insulin-like growth factor 2 |
| chr1_+_201620642 | 20.68 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr10_-_25910298 | 19.05 |
ENSRNOT00000065633
ENSRNOT00000079646 |
Ccng1
|
cyclin G1 |
| chr1_+_167870452 | 14.15 |
ENSRNOT00000025027
|
Olr63
|
olfactory receptor 63 |
| chr9_-_52238564 | 14.04 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chrX_-_136807885 | 14.00 |
ENSRNOT00000010325
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr2_-_105089659 | 12.90 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chrX_-_142248369 | 11.33 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr5_-_7941822 | 11.26 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr19_-_37210412 | 11.22 |
ENSRNOT00000083097
|
B3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr4_+_148782479 | 7.54 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr7_+_42304534 | 6.63 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chrX_+_136460215 | 4.98 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr10_-_44147035 | 3.47 |
ENSRNOT00000041113
|
Olr1424
|
olfactory receptor 1424 |
| chr10_-_43998533 | 3.10 |
ENSRNOT00000047037
|
Olr1417
|
olfactory receptor 1417 |
| chr1_+_40816107 | 2.89 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr15_-_35264841 | 2.67 |
ENSRNOT00000073741
|
LOC100910060
|
granzyme G-like |
| chr7_+_79138925 | 2.20 |
ENSRNOT00000047511
|
AABR07057586.1
|
|
| chr3_-_72171078 | 2.01 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr9_-_44617210 | 1.88 |
ENSRNOT00000074343
|
Lyg1
|
lysozyme G1 |
| chr8_-_19586811 | 1.73 |
ENSRNOT00000091744
|
Olr1149
|
olfactory receptor 1149 |
| chr4_+_72589565 | 1.63 |
ENSRNOT00000079349
|
Olr437
|
olfactory receptor 437 |
| chr7_-_7753978 | 1.19 |
ENSRNOT00000048223
|
Olr1029
|
olfactory receptor 1029 |
| chr5_-_137617258 | 0.80 |
ENSRNOT00000071641
|
Olfr1330-ps1
|
olfactory receptor 1330, pseudogene 1 |
| chr10_+_34402482 | 0.42 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr2_-_140334912 | 0.40 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr17_+_90696019 | 0.04 |
ENSRNOT00000003438
|
Gpr137b
|
G protein-coupled receptor 137B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.7 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 4.0 | 36.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 3.8 | 45.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 3.5 | 14.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 3.2 | 22.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.6 | 11.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.3 | 6.6 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 1.0 | 20.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.7 | 19.0 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.5 | 1.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 2.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 23.0 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.2 | 11.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 2.0 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 11.2 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 23.9 | GO:0010975 | regulation of neuron projection development(GO:0010975) |
| 0.0 | 26.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.0 | 36.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.1 | 20.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 45.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 11.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 25.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 11.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 6.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 43.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 18.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 36.8 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 82.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 3.0 | 20.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 2.0 | 14.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.0 | 22.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.9 | 6.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.9 | 24.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 12.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 25.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 11.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.3 | 14.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 11.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 2.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 18.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 8.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 26.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 22.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 14.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 19.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 20.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 6.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 11.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 22.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 36.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 25.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 14.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 6.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 11.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |