Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxb3
Z-value: 0.57
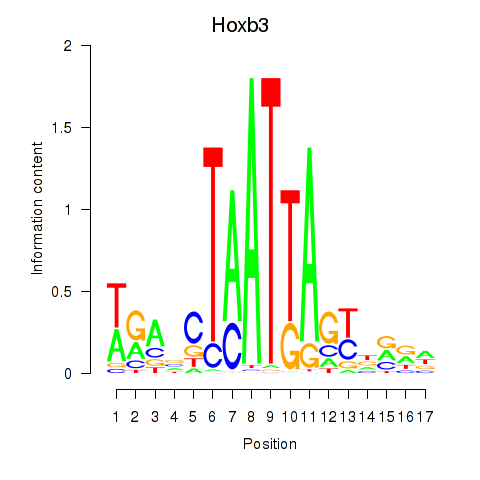
Transcription factors associated with Hoxb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb3
|
ENSRNOG00000008313 | homeo box B3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb3 | rn6_v1_chr10_+_84182118_84182118 | -0.06 | 3.1e-01 | Click! |
Activity profile of Hoxb3 motif
Sorted Z-values of Hoxb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_7058314 | 14.77 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr3_-_14229067 | 13.26 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr2_+_54466280 | 12.82 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr7_-_68549763 | 12.50 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr17_-_43537293 | 9.98 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr9_-_4978892 | 9.23 |
ENSRNOT00000015189
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr2_-_158133861 | 9.10 |
ENSRNOT00000090700
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_23289374 | 9.03 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr14_+_22517774 | 8.80 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_-_104024682 | 8.39 |
ENSRNOT00000056081
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr2_-_191294374 | 8.37 |
ENSRNOT00000067469
|
RGD1562234
|
similar to S100 calcium-binding protein, ventral prostate |
| chr20_-_14020007 | 8.18 |
ENSRNOT00000093521
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_+_61786900 | 7.94 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr17_-_84247038 | 7.17 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr12_-_19167015 | 7.17 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr19_-_50220455 | 7.04 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr2_+_252090669 | 6.97 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr1_-_241460868 | 6.88 |
ENSRNOT00000054776
|
RGD1560242
|
similar to RIKEN cDNA 1700028P14 |
| chr5_-_161981441 | 6.66 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr20_-_9855443 | 6.35 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr1_-_189181901 | 6.32 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr1_-_189182306 | 6.32 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr11_-_61234944 | 5.97 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr7_-_76488216 | 5.91 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr2_+_248398917 | 5.14 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chrX_+_33884499 | 5.01 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr18_-_69944632 | 4.84 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr14_-_7863664 | 4.74 |
ENSRNOT00000061162
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr14_-_16903242 | 4.71 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr4_+_139670092 | 4.62 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr12_-_24775891 | 4.45 |
ENSRNOT00000074851
|
Wbscr27
|
Williams Beuren syndrome chromosome region 27 |
| chr4_+_158088505 | 4.42 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr14_+_22724399 | 4.28 |
ENSRNOT00000002724
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr2_-_60657712 | 4.13 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr1_-_198104109 | 4.11 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr4_-_165192647 | 4.05 |
ENSRNOT00000086461
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr17_+_10137974 | 4.03 |
ENSRNOT00000031935
|
Hk3
|
hexokinase 3 |
| chr8_+_128027958 | 3.89 |
ENSRNOT00000045049
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chrX_-_134866210 | 3.79 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr1_-_252808380 | 3.67 |
ENSRNOT00000025856
|
Ch25h
|
cholesterol 25-hydroxylase |
| chr3_-_79282493 | 3.53 |
ENSRNOT00000049832
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr12_+_24761210 | 3.48 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr9_-_30844199 | 3.33 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr17_+_10138810 | 3.28 |
ENSRNOT00000079850
|
Hk3
|
hexokinase 3 |
| chr14_+_48740190 | 3.27 |
ENSRNOT00000031638
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_-_21920651 | 3.27 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr8_+_13796021 | 3.18 |
ENSRNOT00000013927
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr1_-_189238776 | 3.18 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr9_-_70449796 | 3.11 |
ENSRNOT00000086391
|
Mdh1b
|
malate dehydrogenase 1B |
| chrX_-_6620722 | 3.09 |
ENSRNOT00000066674
|
Maoa
|
monoamine oxidase A |
| chr14_+_76732650 | 3.08 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chrX_-_13279082 | 3.00 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr3_+_128155069 | 2.92 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr7_+_99609191 | 2.86 |
ENSRNOT00000012883
|
Sqle
|
squalene epoxidase |
| chr9_+_98438439 | 2.86 |
ENSRNOT00000027220
|
Scly
|
selenocysteine lyase |
| chr5_-_147412705 | 2.73 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr9_+_20241062 | 2.70 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr4_-_88684415 | 2.67 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr19_-_42180362 | 2.67 |
ENSRNOT00000089515
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr7_+_117456039 | 2.64 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr10_+_103395511 | 2.56 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr1_+_229100752 | 2.48 |
ENSRNOT00000074881
|
LOC686967
|
similar to olfactory receptor 1442 |
| chr1_-_276228574 | 2.46 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr9_+_117795132 | 2.46 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr11_-_81757813 | 2.46 |
ENSRNOT00000002462
|
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr10_+_89484468 | 2.44 |
ENSRNOT00000035523
|
Tmem106a
|
transmembrane protein 106A |
| chr15_+_87722221 | 2.36 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr7_+_59200918 | 2.30 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr14_+_96499520 | 2.27 |
ENSRNOT00000074692
|
AABR07016310.1
|
|
| chr11_+_74057361 | 2.22 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr11_-_14304603 | 2.20 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr4_-_157304653 | 2.15 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr18_+_3705916 | 2.07 |
ENSRNOT00000092846
|
Lama3
|
laminin subunit alpha 3 |
| chr4_+_56981283 | 2.07 |
ENSRNOT00000010989
|
Tspan33
|
tetraspanin 33 |
| chr4_-_165317573 | 2.06 |
ENSRNOT00000087529
ENSRNOT00000080017 |
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr17_+_87274944 | 1.99 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr13_+_90943255 | 1.96 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr17_+_76002275 | 1.94 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr8_-_113937097 | 1.92 |
ENSRNOT00000046816
|
Nek11
|
NIMA-related kinase 11 |
| chr5_+_28485619 | 1.92 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr1_-_184188911 | 1.88 |
ENSRNOT00000055124
ENSRNOT00000014948 |
Calca
|
calcitonin-related polypeptide alpha |
| chr8_+_116318963 | 1.86 |
ENSRNOT00000036556
|
Tusc2
|
tumor suppressor candidate 2 |
| chr10_-_34301197 | 1.76 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr3_-_150603082 | 1.76 |
ENSRNOT00000024310
|
Ahcy
|
adenosylhomocysteinase |
| chr14_-_114692764 | 1.75 |
ENSRNOT00000008210
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_-_34333305 | 1.73 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr9_-_44419998 | 1.72 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr4_+_155009479 | 1.71 |
ENSRNOT00000070906
|
AC127013.1
|
|
| chr18_+_4084228 | 1.63 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr10_-_34361212 | 1.60 |
ENSRNOT00000072557
|
Olr1385
|
olfactory receptor 1385 |
| chr5_-_151459037 | 1.60 |
ENSRNOT00000064472
ENSRNOT00000087836 |
Sytl1
|
synaptotagmin-like 1 |
| chr8_+_2604962 | 1.59 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr16_+_54332660 | 1.55 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr5_+_146656049 | 1.55 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr6_+_104291071 | 1.52 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr18_-_25997555 | 1.52 |
ENSRNOT00000027755
|
Stard4
|
StAR-related lipid transfer domain containing 4 |
| chr5_-_133959447 | 1.46 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr10_+_36517169 | 1.45 |
ENSRNOT00000040534
|
Olr1408
|
olfactory receptor 1408 |
| chr5_-_64850427 | 1.44 |
ENSRNOT00000008883
|
Tmem246
|
transmembrane protein 246 |
| chr10_+_47765432 | 1.43 |
ENSRNOT00000078231
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chr4_-_165026414 | 1.38 |
ENSRNOT00000071421
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr2_+_23385183 | 1.36 |
ENSRNOT00000014860
|
Arsb
|
arylsulfatase B |
| chr1_+_169208988 | 1.34 |
ENSRNOT00000050606
|
Olr140
|
olfactory receptor 140 |
| chr20_-_13994794 | 1.34 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr13_+_83681322 | 1.33 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr4_+_171748273 | 1.32 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr1_+_252284464 | 1.32 |
ENSRNOT00000027969
|
Lipf
|
lipase F, gastric type |
| chr3_-_120076788 | 1.26 |
ENSRNOT00000047158
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr18_+_30581530 | 1.25 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr11_+_81757983 | 1.25 |
ENSRNOT00000088503
|
Tbccd1
|
TBCC domain containing 1 |
| chr10_+_36590325 | 1.25 |
ENSRNOT00000085203
|
Zfp354a
|
zinc finger protein 354A |
| chr4_-_132296413 | 1.24 |
ENSRNOT00000052326
|
RGD1560513
|
similar to macrophage migration inhibitory factor |
| chr10_+_44506098 | 1.24 |
ENSRNOT00000078135
|
LOC501698
|
similar to olfactory receptor Olr1448 |
| chr10_+_96639924 | 1.23 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr20_-_30947484 | 1.22 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr3_+_75970593 | 1.19 |
ENSRNOT00000075834
|
Olr592
|
olfactory receptor 592 |
| chr6_-_105261549 | 1.18 |
ENSRNOT00000009122
|
Synj2bp
|
synaptojanin 2 binding protein |
| chr7_-_44121130 | 1.18 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr16_+_73615949 | 1.16 |
ENSRNOT00000024392
|
Gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr20_-_29199224 | 1.14 |
ENSRNOT00000071477
|
Mcu
|
mitochondrial calcium uniporter |
| chr19_+_653510 | 1.10 |
ENSRNOT00000048221
|
Nae1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr7_-_83670356 | 1.05 |
ENSRNOT00000005584
|
Sybu
|
syntabulin |
| chr20_+_31761405 | 1.05 |
ENSRNOT00000000661
|
Neurog3
|
neurogenin 3 |
| chr17_+_9797907 | 1.05 |
ENSRNOT00000021638
|
Lman2
|
lectin, mannose-binding 2 |
| chr4_+_148476943 | 1.01 |
ENSRNOT00000017689
|
Olr823
|
olfactory receptor 823 |
| chr16_-_29936307 | 1.01 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr10_-_88670430 | 0.99 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr1_+_190200324 | 0.95 |
ENSRNOT00000001933
|
Abca16
|
ATP-binding cassette, subfamily A (ABC1), member 16 |
| chr14_-_45859908 | 0.95 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr1_-_227457629 | 0.94 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr14_-_2032593 | 0.94 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr6_-_3355339 | 0.94 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr6_-_108920767 | 0.90 |
ENSRNOT00000006704
|
Prox2
|
prospero homeobox 2 |
| chr14_+_2892753 | 0.89 |
ENSRNOT00000061630
|
Evi5
|
ecotropic viral integration site 5 |
| chr4_+_71836677 | 0.88 |
ENSRNOT00000024048
|
Tas2r126
|
taste receptor, type 2, member 126 |
| chr20_-_1099336 | 0.87 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chr3_-_77494108 | 0.86 |
ENSRNOT00000047726
|
Olr662
|
olfactory receptor 662 |
| chr2_-_46544457 | 0.86 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr10_+_34402482 | 0.86 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr10_-_60032218 | 0.85 |
ENSRNOT00000072328
|
Olr1481
|
olfactory receptor 1481 |
| chr17_+_81280898 | 0.85 |
ENSRNOT00000057785
|
Tmem236
|
transmembrane protein 236 |
| chr16_-_75241303 | 0.84 |
ENSRNOT00000058056
|
Defb2
|
defensin beta 2 |
| chr2_+_85305225 | 0.80 |
ENSRNOT00000015904
|
Tas2r119
|
taste receptor, type 2, member 119 |
| chr5_-_151473750 | 0.78 |
ENSRNOT00000011357
|
Tmem222
|
transmembrane protein 222 |
| chr1_-_78686952 | 0.77 |
ENSRNOT00000047079
|
LOC679748
|
similar to Macrophage migration inhibitory factor (MIF) (Phenylpyruvate tautomerase) (Glycosylation-inhibiting factor) (GIF) (Delayed early response protein 6) (DER6) |
| chr2_+_148765412 | 0.75 |
ENSRNOT00000018248
ENSRNOT00000084662 |
Selenot
|
selenoprotein T |
| chr12_-_35979193 | 0.75 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr7_-_83701447 | 0.75 |
ENSRNOT00000088893
|
AABR07057675.1
|
|
| chr10_-_44471915 | 0.73 |
ENSRNOT00000089901
|
Olr1440
|
olfactory receptor 1440 |
| chr8_+_77088469 | 0.73 |
ENSRNOT00000090317
|
Gm10642
|
predicted gene 10642 |
| chr8_-_59239954 | 0.72 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr1_-_167971151 | 0.72 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr4_-_174180729 | 0.72 |
ENSRNOT00000011376
|
Plcz1
|
phospholipase C, zeta 1 |
| chr13_+_82438697 | 0.71 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr2_-_187786700 | 0.71 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr10_+_5199226 | 0.70 |
ENSRNOT00000003544
|
Dexi
|
dexamethasone-induced transcript |
| chr8_+_128792707 | 0.70 |
ENSRNOT00000083329
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr12_+_22727335 | 0.69 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr17_-_22678253 | 0.69 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr1_-_149675443 | 0.68 |
ENSRNOT00000086084
|
Olr294
|
olfactory receptor 294 |
| chr1_+_169153297 | 0.68 |
ENSRNOT00000052078
|
Olr137
|
olfactory receptor 137 |
| chr20_-_3344286 | 0.68 |
ENSRNOT00000091505
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr5_+_14890408 | 0.67 |
ENSRNOT00000033116
|
Sox17
|
SRY box 17 |
| chr11_+_43465370 | 0.67 |
ENSRNOT00000039939
|
Olr1547
|
olfactory receptor 1547 |
| chr4_+_52235811 | 0.65 |
ENSRNOT00000060478
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr4_+_148782479 | 0.65 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr3_-_44501115 | 0.65 |
ENSRNOT00000087071
|
Acvr1
|
activin A receptor type 1 |
| chr1_+_169182969 | 0.63 |
ENSRNOT00000040131
|
Olr139
|
olfactory receptor 139 |
| chr5_+_187312 | 0.62 |
ENSRNOT00000030857
|
LOC102557117
|
zinc finger protein 120-like |
| chr6_+_104291340 | 0.59 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr10_+_44679832 | 0.59 |
ENSRNOT00000044332
|
Olr1450
|
olfactory receptor 1450 |
| chr16_-_75107931 | 0.58 |
ENSRNOT00000058066
|
Defb15
|
defensin beta 15 |
| chr2_+_185846232 | 0.58 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr6_-_103807838 | 0.57 |
ENSRNOT00000058328
|
AABR07064998.1
|
|
| chr18_+_16544508 | 0.56 |
ENSRNOT00000020601
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr20_-_36061477 | 0.56 |
ENSRNOT00000038249
|
RGD1559810
|
similar to hypothetical protein |
| chr1_-_150101177 | 0.55 |
ENSRNOT00000042931
|
Olr24
|
olfactory receptor 24 |
| chr3_-_164964702 | 0.55 |
ENSRNOT00000014595
|
Adnp
|
activity-dependent neuroprotector homeobox |
| chr11_-_1437732 | 0.52 |
ENSRNOT00000037345
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr1_-_64147251 | 0.51 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr20_+_44680449 | 0.50 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr4_-_82141385 | 0.50 |
ENSRNOT00000008447
|
Hoxa3
|
homeobox A3 |
| chrX_+_14019961 | 0.49 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr1_-_14117021 | 0.49 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr13_+_70174936 | 0.48 |
ENSRNOT00000064068
ENSRNOT00000079861 ENSRNOT00000092562 |
Arpc5
|
actin related protein 2/3 complex, subunit 5 |
| chr15_+_33885106 | 0.47 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr10_-_41491554 | 0.47 |
ENSRNOT00000035759
|
LOC102552619
|
uncharacterized LOC102552619 |
| chr13_+_56096834 | 0.47 |
ENSRNOT00000035129
|
Dennd1b
|
DENN domain containing 1B |
| chr17_+_11856525 | 0.45 |
ENSRNOT00000014546
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr1_-_167732783 | 0.43 |
ENSRNOT00000024965
|
Olr44
|
olfactory receptor 44 |
| chr6_-_108660063 | 0.43 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr3_-_102672294 | 0.41 |
ENSRNOT00000030909
|
Olr760
|
olfactory receptor 760 |
| chr10_+_44650570 | 0.40 |
ENSRNOT00000047671
|
Olr1449
|
olfactory receptor 1449 |
| chr1_+_75326217 | 0.40 |
ENSRNOT00000044964
|
Vom1r62
|
vomeronasal 1 receptor 62 |
| chr5_-_59266293 | 0.40 |
ENSRNOT00000050490
|
Olr836
|
olfactory receptor 836 |
| chr10_-_5344121 | 0.40 |
ENSRNOT00000003479
|
Nubp1
|
nucleotide binding protein 1 |
| chr20_-_35342 | 0.40 |
ENSRNOT00000048277
|
LOC100910263
|
olfactory receptor 2B11-like |
| chr3_-_103152122 | 0.40 |
ENSRNOT00000045743
|
Olr781
|
olfactory receptor 781 |
| chr1_-_198450047 | 0.39 |
ENSRNOT00000027360
|
Mvp
|
major vault protein |
| chr8_-_14156656 | 0.38 |
ENSRNOT00000089364
|
Deup1
|
deuterosome assembly protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.8 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 3.7 | 14.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 3.2 | 12.6 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 3.0 | 9.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 2.8 | 13.8 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 2.4 | 7.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 2.2 | 13.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 2.0 | 8.2 | GO:0031179 | peptide modification(GO:0031179) |
| 1.1 | 10.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 1.0 | 2.9 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) regulation of monocyte extravasation(GO:2000437) |
| 0.9 | 3.8 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.9 | 10.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.9 | 5.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.6 | 1.9 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.6 | 1.9 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.6 | 7.3 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.6 | 4.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.6 | 2.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.5 | 4.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.4 | 3.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.4 | 7.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 2.9 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.4 | 1.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 1.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.4 | 1.5 | GO:0070859 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.4 | 4.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 3.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.3 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 6.0 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.3 | 1.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.3 | 1.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.2 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.2 | 3.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 0.7 | GO:0061010 | inner cell mass cellular morphogenesis(GO:0001828) endodermal cell fate determination(GO:0007493) gall bladder development(GO:0061010) |
| 0.2 | 1.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 0.6 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.2 | 1.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 2.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 0.5 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.2 | 1.8 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.2 | 0.5 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.9 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.7 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 0.1 | 0.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 3.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.9 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 2.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.0 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.1 | 0.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.4 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.7 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.8 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 2.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.5 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.5 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) gas homeostasis(GO:0033483) |
| 0.1 | 1.4 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 4.7 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 1.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 2.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 2.2 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 2.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 7.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 4.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.8 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 5.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 6.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.4 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 4.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 1.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 3.5 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 2.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 1.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 2.1 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 0.9 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 3.4 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 26.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.3 | 5.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.7 | 4.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 2.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.3 | 7.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 5.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 1.6 | GO:0072558 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 2.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.8 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.1 | 3.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 1.1 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 3.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 6.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 7.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 13.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 4.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 7.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 13.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 10.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 4.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 3.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.6 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 4.1 | 4.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 2.8 | 13.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 2.3 | 9.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 1.7 | 7.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.6 | 9.5 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 1.3 | 5.1 | GO:0019002 | GMP binding(GO:0019002) |
| 1.2 | 7.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.0 | 10.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 0.9 | 9.0 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.8 | 6.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.8 | 3.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.6 | 3.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.5 | 14.8 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.5 | 3.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 1.8 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 13.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 1.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 7.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 1.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.3 | 1.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 3.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 5.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 3.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 5.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 0.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.2 | 0.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 4.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 3.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 3.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 4.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 12.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 3.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 2.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 2.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.5 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 3.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 2.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 4.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 7.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 7.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.9 | 12.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 26.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 5.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 9.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 4.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 4.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 2.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 4.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 7.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 8.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |