Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxb2_Dlx2
Z-value: 0.54
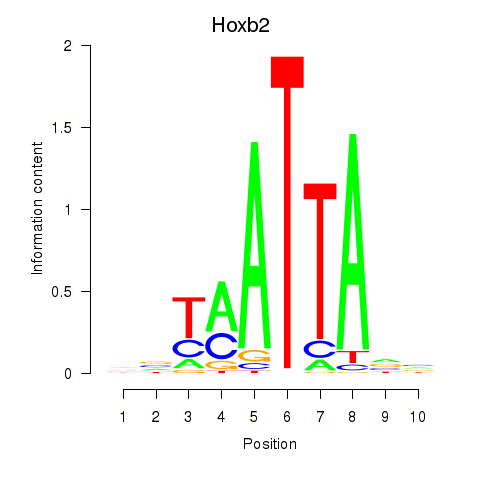
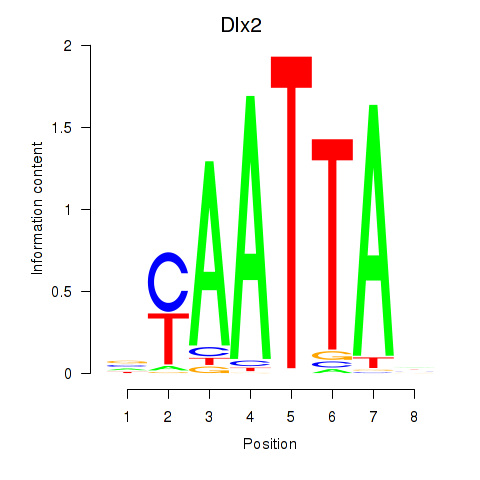
Transcription factors associated with Hoxb2_Dlx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb2
|
ENSRNOG00000008365 | homeobox B2 |
|
Dlx2
|
ENSRNOG00000001519 | distal-less homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx2 | rn6_v1_chr3_-_58181971_58181971 | 0.35 | 1.3e-10 | Click! |
| Hoxb2 | rn6_v1_chr10_+_84200880_84200880 | 0.17 | 1.7e-03 | Click! |
Activity profile of Hoxb2_Dlx2 motif
Sorted Z-values of Hoxb2_Dlx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_84064427 | 22.86 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr5_+_33784715 | 20.59 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr8_+_33239139 | 19.56 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr9_-_30844199 | 18.23 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr8_-_78233430 | 17.15 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr1_+_198383201 | 16.44 |
ENSRNOT00000037405
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr4_-_17594598 | 16.27 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr6_+_58468155 | 12.98 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr1_-_279277339 | 12.97 |
ENSRNOT00000023667
|
Gfra1
|
GDNF family receptor alpha 1 |
| chr11_+_58624198 | 12.32 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr18_-_26656879 | 11.05 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr16_+_2706428 | 10.87 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr17_+_47721977 | 10.80 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr2_-_158156444 | 10.48 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_+_50049828 | 10.04 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chrX_+_118197217 | 9.64 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr8_-_14880644 | 9.40 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr17_-_9762813 | 9.35 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr17_+_11683862 | 9.07 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chrX_-_32095867 | 8.72 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr1_+_13595295 | 8.69 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr2_-_35104963 | 8.41 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr8_-_45137893 | 8.30 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr2_-_158156150 | 8.13 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_23289374 | 7.92 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr7_+_78558701 | 7.90 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr3_-_66417741 | 7.74 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr16_+_46731403 | 7.61 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr18_-_31343797 | 7.48 |
ENSRNOT00000081850
|
Pcdh1
|
protocadherin 1 |
| chr12_-_2174131 | 7.48 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr12_-_29743705 | 7.44 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr8_-_87419564 | 7.42 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr16_-_39476025 | 7.37 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr2_+_18392142 | 7.23 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_+_87224677 | 7.11 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_-_215033460 | 7.07 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr18_-_6781841 | 6.85 |
ENSRNOT00000077606
ENSRNOT00000048109 |
Aqp4
|
aquaporin 4 |
| chr11_-_782954 | 6.32 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr4_+_2055615 | 6.27 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr5_-_93244202 | 6.20 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr2_-_261337163 | 5.98 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chr13_+_70157522 | 5.88 |
ENSRNOT00000036906
|
Apobec4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr16_+_23553647 | 5.84 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr10_-_87248572 | 5.82 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr7_-_76488216 | 5.77 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr12_-_10335499 | 5.76 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr18_+_30592794 | 5.56 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr14_-_19072677 | 5.56 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr2_+_18354542 | 5.54 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chrX_-_40086870 | 5.43 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr13_+_57243877 | 5.42 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chrX_+_33895769 | 5.42 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr10_-_91661558 | 5.37 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr13_-_76049363 | 5.23 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr4_-_55011415 | 5.17 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr20_-_25826658 | 5.13 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr3_-_51612397 | 5.10 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr5_+_6373583 | 5.03 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr3_+_48096954 | 5.00 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr17_+_63635086 | 5.00 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr5_+_58855773 | 4.99 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr2_-_236480502 | 4.86 |
ENSRNOT00000015020
|
Sgms2
|
sphingomyelin synthase 2 |
| chr1_+_44311513 | 4.85 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr4_+_158088505 | 4.75 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr7_+_42269784 | 4.75 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr11_+_45751812 | 4.74 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr15_+_106434177 | 4.69 |
ENSRNOT00000015271
|
Farp1
|
FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| chr20_+_25990656 | 4.68 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chrM_+_7006 | 4.63 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr3_+_117421604 | 4.63 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr2_+_266315036 | 4.61 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr3_+_45683993 | 4.61 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_+_22950018 | 4.58 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr4_+_172942020 | 4.54 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr16_+_54332660 | 4.45 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chrX_+_110789269 | 4.32 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr3_+_128828331 | 4.25 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr19_+_53044379 | 4.22 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr4_-_89695928 | 4.19 |
ENSRNOT00000039316
|
Gprin3
|
GPRIN family member 3 |
| chr2_-_5577369 | 4.16 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr15_+_11298478 | 4.13 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr3_-_14229067 | 4.09 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr3_-_148932878 | 4.01 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr9_-_85243001 | 3.99 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr9_+_73378057 | 3.99 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr7_+_42304534 | 3.90 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr8_+_33514042 | 3.89 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr17_-_84247038 | 3.87 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr6_-_71199110 | 3.86 |
ENSRNOT00000081883
|
Prkd1
|
protein kinase D1 |
| chr18_+_30435119 | 3.84 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr3_+_151335292 | 3.84 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr19_-_58735173 | 3.82 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chrM_+_11736 | 3.79 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr8_-_53816447 | 3.77 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr14_+_37130743 | 3.75 |
ENSRNOT00000031756
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr18_+_30474947 | 3.72 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr5_-_164714145 | 3.72 |
ENSRNOT00000055680
|
LOC100911485
|
mitofusin-2-like |
| chr4_-_64330996 | 3.70 |
ENSRNOT00000016088
|
Ptn
|
pleiotrophin |
| chr18_+_30509393 | 3.67 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr18_+_81821127 | 3.67 |
ENSRNOT00000058199
|
Fbxo15
|
F-box protein 15 |
| chr9_+_95501778 | 3.64 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr11_+_24266345 | 3.60 |
ENSRNOT00000046973
|
Jam2
|
junctional adhesion molecule 2 |
| chr1_-_67065797 | 3.57 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr7_-_69982592 | 3.56 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr3_+_48106099 | 3.50 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr19_+_25043680 | 3.50 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr4_+_94696965 | 3.40 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr7_-_107203897 | 3.30 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr4_-_82173207 | 3.24 |
ENSRNOT00000074167
|
Hoxa5
|
homeo box A5 |
| chr3_+_140024043 | 3.22 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr1_+_242572533 | 3.21 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr1_-_250626844 | 3.19 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr5_-_28130803 | 3.16 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr9_+_73418607 | 3.13 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr5_-_102743417 | 3.12 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr3_-_161917285 | 3.10 |
ENSRNOT00000025150
|
Cdh22
|
cadherin 22 |
| chr9_-_44419998 | 3.07 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr2_-_57935334 | 3.04 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr6_-_3355339 | 3.02 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chrM_+_9451 | 3.00 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr9_+_81566074 | 2.99 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chrM_+_9870 | 2.98 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr1_-_48825364 | 2.97 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr6_-_114476723 | 2.93 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr15_-_20783063 | 2.92 |
ENSRNOT00000083268
|
Bmp4
|
bone morphogenetic protein 4 |
| chr7_-_52404774 | 2.89 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr1_+_70260041 | 2.88 |
ENSRNOT00000020416
|
Zim1
|
zinc finger, imprinted 1 |
| chr9_+_95398237 | 2.88 |
ENSRNOT00000025879
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr10_-_91986632 | 2.82 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr10_-_16731898 | 2.81 |
ENSRNOT00000028186
|
Crebrf
|
CREB3 regulatory factor |
| chr10_+_90230711 | 2.79 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr9_+_73493027 | 2.78 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chr1_+_185210922 | 2.72 |
ENSRNOT00000055120
|
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr2_-_30634243 | 2.72 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr8_-_17524839 | 2.70 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr2_+_145174876 | 2.68 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr1_-_173764246 | 2.67 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr3_-_77227036 | 2.66 |
ENSRNOT00000072638
|
LOC103691843
|
olfactory receptor 4P4-like |
| chr3_+_14889510 | 2.62 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr2_-_144467912 | 2.59 |
ENSRNOT00000040002
|
Ccna1
|
cyclin A1 |
| chr11_-_28478360 | 2.57 |
ENSRNOT00000032663
|
Cldn17
|
claudin 17 |
| chr2_+_136993208 | 2.57 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr5_-_117754571 | 2.56 |
ENSRNOT00000011405
|
Dock7
|
dedicator of cytokinesis 7 |
| chr20_+_1749716 | 2.56 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr5_-_28131133 | 2.55 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr14_+_37116492 | 2.55 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr18_-_31109025 | 2.53 |
ENSRNOT00000026381
|
Fchsd1
|
FCH and double SH3 domains 1 |
| chr6_+_73553210 | 2.52 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr5_-_150704117 | 2.47 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr4_-_150485781 | 2.45 |
ENSRNOT00000008763
|
Zfp248
|
zinc finger protein 248 |
| chr2_+_206314213 | 2.43 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr6_-_7058314 | 2.43 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr8_+_106449321 | 2.42 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr4_+_25686392 | 2.39 |
ENSRNOT00000061199
|
Cldn12
|
claudin 12 |
| chr2_+_225645568 | 2.38 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr7_-_68549763 | 2.35 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr12_-_5682608 | 2.33 |
ENSRNOT00000076483
|
Fry
|
FRY microtubule binding protein |
| chr3_-_102672294 | 2.33 |
ENSRNOT00000030909
|
Olr760
|
olfactory receptor 760 |
| chr3_+_95715193 | 2.33 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr2_+_226563050 | 2.32 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr2_-_96509424 | 2.32 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr4_+_68656928 | 2.32 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr7_+_44009069 | 2.31 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr12_+_30606161 | 2.30 |
ENSRNOT00000093601
|
Sept14
|
septin 14 |
| chr2_-_33025271 | 2.30 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_52658275 | 2.28 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr15_+_56666012 | 2.28 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr2_+_66940057 | 2.22 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr5_-_28164326 | 2.19 |
ENSRNOT00000088165
|
Slc26a7
|
solute carrier family 26 member 7 |
| chrX_+_111735820 | 2.19 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr2_-_188559882 | 2.15 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr1_+_87563975 | 2.14 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chrX_+_159158194 | 2.09 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr14_-_16903242 | 2.07 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr12_+_41486076 | 2.03 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chrX_+_65040775 | 2.02 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chrM_+_10160 | 2.01 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr2_+_72006099 | 2.00 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr1_-_261371508 | 1.99 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr6_+_101532518 | 1.99 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr2_+_144861455 | 1.97 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr2_+_51672722 | 1.95 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr2_-_46544457 | 1.93 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr4_-_135069970 | 1.92 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr5_-_12526962 | 1.91 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr6_+_106496992 | 1.91 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr8_-_39551700 | 1.91 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr3_-_57646572 | 1.91 |
ENSRNOT00000076202
|
Mettl8
|
methyltransferase like 8 |
| chr6_+_28382962 | 1.91 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr19_+_39229754 | 1.91 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr4_-_176909075 | 1.89 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chrX_+_65040934 | 1.89 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr14_-_21128505 | 1.85 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr3_+_149643481 | 1.84 |
ENSRNOT00000020713
|
Bpifa5
|
BPI fold containing family A, member 5 |
| chr8_-_104593625 | 1.82 |
ENSRNOT00000016625
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr5_-_130085838 | 1.81 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr8_-_128754514 | 1.81 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr14_-_72380330 | 1.80 |
ENSRNOT00000082653
ENSRNOT00000081878 ENSRNOT00000006727 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr5_-_117612123 | 1.79 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr2_+_248398917 | 1.79 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb2_Dlx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 11.9 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 3.1 | 9.4 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 3.0 | 9.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 2.9 | 8.7 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 2.6 | 7.9 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 2.1 | 6.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 1.9 | 3.9 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 1.7 | 8.6 | GO:0070668 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 1.7 | 6.8 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 1.5 | 7.7 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.5 | 4.6 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 1.5 | 4.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.3 | 5.2 | GO:0060435 | bronchiole development(GO:0060435) |
| 1.3 | 3.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.2 | 3.7 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 1.2 | 12.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.1 | 13.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.1 | 4.2 | GO:0072011 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 1.0 | 3.0 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 1.0 | 2.9 | GO:0072098 | intermediate mesoderm development(GO:0048389) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) ureter urothelium development(GO:0072190) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) cardiac jelly development(GO:1905072) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.9 | 2.8 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.9 | 2.7 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.9 | 16.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.8 | 1.7 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.8 | 2.5 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.8 | 2.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.8 | 2.4 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.8 | 2.3 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.8 | 3.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.8 | 3.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.8 | 6.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.7 | 5.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.7 | 2.8 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.7 | 1.4 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.7 | 4.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.7 | 2.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.7 | 2.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.7 | 2.6 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.6 | 7.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.6 | 5.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.6 | 1.9 | GO:0036258 | multivesicular body assembly(GO:0036258) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.6 | 2.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.6 | 4.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 6.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.6 | 7.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.6 | 1.8 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.6 | 3.0 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.6 | 4.8 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.6 | 2.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.6 | 2.9 | GO:0050955 | thermoception(GO:0050955) |
| 0.6 | 9.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.6 | 4.5 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.5 | 6.0 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.5 | 5.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.5 | 16.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 1.9 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.5 | 2.3 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 1.8 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.4 | 1.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.4 | 17.0 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.4 | 1.3 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.4 | 1.8 | GO:0002933 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.4 | 0.9 | GO:0060769 | embryonic skeletal limb joint morphogenesis(GO:0036023) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.4 | 4.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 1.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.4 | 4.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.4 | 1.9 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.4 | 13.9 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.4 | 3.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 2.9 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.4 | 9.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 0.7 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.3 | 6.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.3 | 1.7 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.3 | 3.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.3 | 3.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.3 | 2.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.3 | 2.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 3.8 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.3 | 3.4 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.9 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.3 | 3.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 4.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 5.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 4.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 0.9 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 3.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.7 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 1.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 3.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 5.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 2.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 13.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 7.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 4.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.2 | 22.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 4.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 0.5 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.2 | 1.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 3.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 1.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 2.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 1.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 3.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 6.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 1.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 3.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 12.9 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.1 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 5.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 14.0 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 2.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 5.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 2.1 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 2.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 2.7 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.3 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 30.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 4.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 3.8 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 0.5 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.9 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.9 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 1.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 4.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 1.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 1.8 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0042753 | negative regulation of NF-kappaB import into nucleus(GO:0042347) positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.8 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 1.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 3.2 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 4.4 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.9 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 5.8 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 2.0 | GO:0051168 | nuclear export(GO:0051168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 15.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.6 | 4.9 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 1.5 | 12.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.1 | 5.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.9 | 2.6 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.8 | 2.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.8 | 4.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.6 | 3.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 2.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.5 | 1.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.4 | 1.8 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.4 | 2.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 2.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 6.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 2.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 0.9 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.3 | 4.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 1.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 0.8 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 8.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 4.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 7.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 5.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 9.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 17.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 11.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 7.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 1.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 8.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 2.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 1.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 5.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 3.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 6.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 5.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 13.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 4.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 16.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 9.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 7.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 8.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 6.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 3.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 3.4 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 0.9 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 5.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 7.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 4.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 16.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 5.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 20.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 4.8 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 29.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 5.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 3.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 8.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 7.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.7 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 11.9 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 2.8 | 11.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 2.2 | 8.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.9 | 9.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.5 | 4.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 1.4 | 4.1 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 1.4 | 10.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.3 | 5.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.3 | 16.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.2 | 8.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.2 | 3.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.0 | 6.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 1.0 | 3.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.0 | 4.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.9 | 4.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.7 | 4.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.6 | 6.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.6 | 1.9 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.6 | 3.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.6 | 3.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.6 | 5.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 1.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 3.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 7.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 4.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 3.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 2.0 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.5 | 7.9 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.5 | 6.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 1.9 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.5 | 2.4 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.5 | 2.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 12.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 1.8 | GO:0019002 | GMP binding(GO:0019002) |
| 0.4 | 2.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.4 | 11.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 2.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 2.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 1.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 1.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 1.4 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.3 | 2.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 5.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 1.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 8.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 2.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 6.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 3.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 3.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 7.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 3.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 2.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 5.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 5.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.8 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.2 | 4.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 3.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 4.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 13.6 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.2 | 1.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 5.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 5.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 3.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 5.9 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.2 | 0.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 21.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 7.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 1.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 2.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 0.5 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 3.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.2 | 4.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 3.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 4.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 4.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 34.2 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 12.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 9.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 2.4 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.1 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 8.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 6.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 3.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 39.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 7.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 5.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 4.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 2.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 18.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 13.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 12.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 3.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 19.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 14.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 6.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 6.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 8.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 7.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 2.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 4.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 7.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 5.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 3.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 4.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 13.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 0.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 3.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 16.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.9 | 11.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.6 | 6.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 13.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 4.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 5.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 8.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.3 | 9.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 18.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 2.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 9.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 5.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 5.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 5.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 2.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 2.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 2.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 2.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 4.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 1.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 4.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 5.8 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 3.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 3.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 3.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |