Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
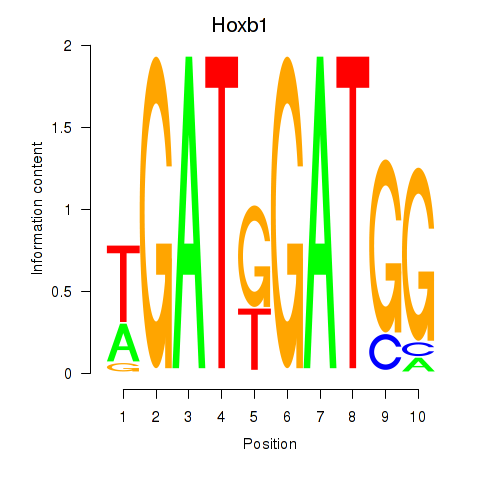
Results for Hoxb1
Z-value: 0.57
Transcription factors associated with Hoxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb1
|
ENSRNOG00000008506 | homeo box B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb1 | rn6_v1_chr10_+_84214358_84214358 | 0.03 | 5.4e-01 | Click! |
Activity profile of Hoxb1 motif
Sorted Z-values of Hoxb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_8498422 | 21.94 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr3_+_51883559 | 20.81 |
ENSRNOT00000007197
|
Csrnp3
|
cysteine and serine rich nuclear protein 3 |
| chr10_+_86340940 | 20.47 |
ENSRNOT00000073486
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr7_-_29171783 | 19.90 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr10_-_85974644 | 18.69 |
ENSRNOT00000006098
ENSRNOT00000082974 |
Cacnb1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr1_-_215838209 | 18.66 |
ENSRNOT00000050760
|
Igf2
|
insulin-like growth factor 2 |
| chr1_+_25173242 | 18.33 |
ENSRNOT00000016518
|
Clvs2
|
clavesin 2 |
| chr5_-_109651730 | 15.85 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr10_+_5930298 | 15.54 |
ENSRNOT00000044626
|
Grin2a
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr18_+_61563053 | 14.41 |
ENSRNOT00000022845
|
Grp
|
gastrin releasing peptide |
| chr6_+_58468155 | 14.16 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr5_+_124021366 | 13.97 |
ENSRNOT00000009977
|
Dab1
|
DAB1, reelin adaptor protein |
| chr16_+_46731403 | 12.16 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_260992291 | 11.96 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr4_-_155275161 | 10.68 |
ENSRNOT00000032690
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr10_-_25910298 | 10.54 |
ENSRNOT00000065633
ENSRNOT00000079646 |
Ccng1
|
cyclin G1 |
| chr5_-_12526962 | 10.27 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr15_-_33285779 | 9.03 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chrX_+_82143789 | 8.56 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr6_+_26387877 | 7.54 |
ENSRNOT00000076105
|
Fndc4
|
|
| chr19_-_43290363 | 7.52 |
ENSRNOT00000024248
|
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr3_-_23020441 | 7.35 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr19_-_37427989 | 6.90 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_+_17602281 | 5.96 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr1_-_141470380 | 5.79 |
ENSRNOT00000065759
|
Plin1
|
perilipin 1 |
| chr13_+_71107465 | 5.46 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr7_+_124680294 | 4.98 |
ENSRNOT00000014720
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr19_-_24614019 | 4.60 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr3_+_116899878 | 4.07 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr7_+_27089278 | 3.59 |
ENSRNOT00000082398
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr8_+_13796021 | 3.11 |
ENSRNOT00000013927
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr16_-_74864816 | 2.48 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr6_+_29061618 | 1.97 |
ENSRNOT00000092223
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr8_-_82339937 | 1.82 |
ENSRNOT00000080797
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr16_-_71203609 | 1.78 |
ENSRNOT00000088458
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr1_+_40816107 | 1.59 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr10_-_63499390 | 1.56 |
ENSRNOT00000032934
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr6_+_102392828 | 1.54 |
ENSRNOT00000089162
|
Rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr2_-_184993341 | 1.46 |
ENSRNOT00000071580
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr11_-_38457373 | 1.35 |
ENSRNOT00000041177
|
Zfp295
|
zinc finger protein 295 |
| chr1_+_229443102 | 0.93 |
ENSRNOT00000067320
|
Olr334
|
olfactory receptor 334 |
| chr14_+_3506339 | 0.84 |
ENSRNOT00000002867
|
Tgfbr3
|
transforming growth factor beta receptor 3 |
| chr12_-_22748860 | 0.73 |
ENSRNOT00000001923
|
Cldn15
|
claudin 15 |
| chr15_-_8914501 | 0.46 |
ENSRNOT00000008752
|
Thrb
|
thyroid hormone receptor beta |
| chr1_-_169973241 | 0.35 |
ENSRNOT00000023315
|
Olr191-ps
|
olfactory receptor pseudogene 191 |
| chr6_+_8218696 | 0.16 |
ENSRNOT00000083470
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr3_+_141225290 | 0.10 |
ENSRNOT00000016351
|
Xrn2
|
5'-3' exoribonuclease 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0033058 | directional locomotion(GO:0033058) |
| 4.7 | 14.0 | GO:0021812 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 3.4 | 20.5 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 3.0 | 12.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 2.7 | 18.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.0 | 19.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.8 | 7.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 1.2 | 14.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.1 | 12.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.0 | 21.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.6 | 8.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.5 | 4.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 18.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.4 | 5.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.4 | 10.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.3 | 7.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 18.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.3 | 0.8 | GO:1902338 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of apoptotic process involved in morphogenesis(GO:1902338) negative regulation of apoptotic process involved in development(GO:1904746) |
| 0.2 | 20.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 2.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 2.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 1.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 5.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 4.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 14.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 6.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 0.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.1 | 9.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 1.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 1.8 | GO:0034968 | histone lysine methylation(GO:0034968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 19.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.3 | 15.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.0 | 20.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.5 | 3.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 18.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 0.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 18.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 7.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 5.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 14.0 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 5.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 14.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 7.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 32.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 10.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 6.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 1.9 | 7.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 1.7 | 12.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.6 | 15.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.5 | 19.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.0 | 18.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.9 | 8.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 18.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.8 | 2.5 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.8 | 18.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.7 | 9.0 | GO:0045294 | alpha-catenin binding(GO:0045294) delta-catenin binding(GO:0070097) |
| 0.5 | 14.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 4.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 14.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 1.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 20.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 5.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 7.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 21.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.8 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 13.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 13.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 6.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 10.5 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 7.4 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 14.9 | GO:0003723 | RNA binding(GO:0003723) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 29.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.5 | 14.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 30.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 10.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 20.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.9 | 18.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 15.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.7 | 14.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.5 | 19.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 7.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 5.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 16.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 9.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 4.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 12.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 7.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 5.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |