Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxa7_Hoxc8
Z-value: 0.77
Transcription factors associated with Hoxa7_Hoxc8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
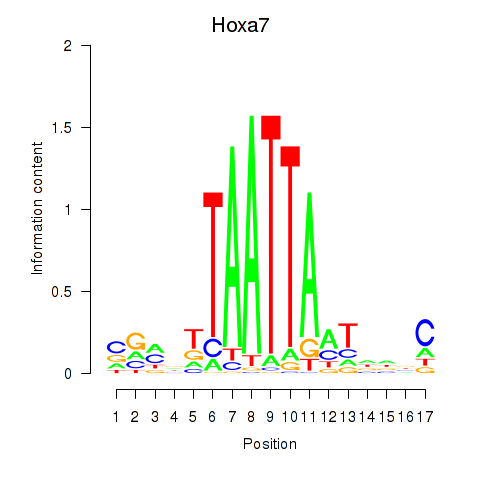
Hoxa7
|
ENSRNOG00000049858 | homeobox A7 |
|
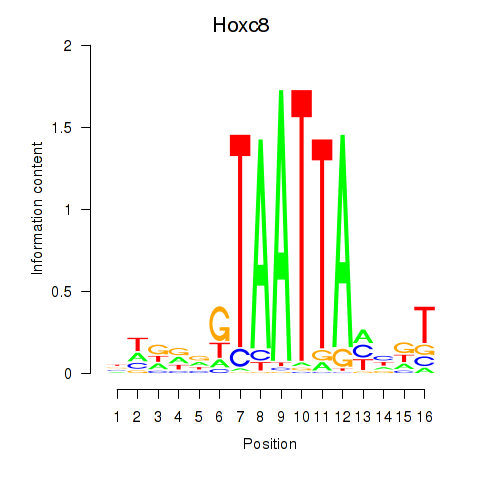
Hoxc8
|
ENSRNOG00000028619 | homeobox C8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc8 | rn6_v1_chr7_+_144605058_144605058 | 0.63 | 6.1e-37 | Click! |
| Hoxa7 | rn6_v1_chr4_-_82271893_82271893 | 0.16 | 4.9e-03 | Click! |
Activity profile of Hoxa7_Hoxc8 motif
Sorted Z-values of Hoxa7_Hoxc8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_64789940 | 63.79 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr3_-_15278645 | 27.43 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr9_+_73378057 | 26.09 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr2_-_57935334 | 21.75 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr9_+_73418607 | 19.16 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_236900904 | 18.56 |
ENSRNOT00000066846
|
AABR07006480.1
|
|
| chr14_+_39964588 | 17.91 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chrX_+_84064427 | 17.77 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr2_+_145174876 | 16.97 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr1_-_236729412 | 16.31 |
ENSRNOT00000054794
|
Prune2
|
prune homolog 2 |
| chr8_+_33239139 | 15.73 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr15_+_62406873 | 15.55 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr9_+_67234303 | 13.76 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr4_+_94696965 | 12.58 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr2_-_33025271 | 12.13 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_170205591 | 11.94 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chrX_+_14019961 | 10.05 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr3_+_48096954 | 9.27 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr18_+_30036887 | 8.61 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr2_-_35870578 | 8.48 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chrX_+_131381134 | 7.92 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr12_-_52658275 | 7.88 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr4_-_150485781 | 7.70 |
ENSRNOT00000008763
|
Zfp248
|
zinc finger protein 248 |
| chr10_-_74679858 | 7.55 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr8_-_128754514 | 7.31 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr8_-_17696989 | 6.80 |
ENSRNOT00000083752
|
AABR07069336.1
|
|
| chr18_+_14757679 | 6.53 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_-_52849907 | 6.51 |
ENSRNOT00000041096
|
Scn7a
|
sodium voltage-gated channel alpha subunit 7 |
| chr2_+_72006099 | 6.47 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr18_+_30592794 | 6.29 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr8_+_82038967 | 6.28 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr10_+_84135116 | 6.24 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr9_+_84203072 | 6.19 |
ENSRNOT00000018882
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr5_-_133959447 | 6.07 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr4_+_148208260 | 5.95 |
ENSRNOT00000015742
|
Zfand4
|
zinc finger AN1-type containing 4 |
| chr9_+_71915421 | 5.80 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr5_+_58661049 | 5.73 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr2_-_98610368 | 5.69 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr4_-_18396035 | 5.67 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chrX_-_69218526 | 5.60 |
ENSRNOT00000092321
ENSRNOT00000074071 ENSRNOT00000092571 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr3_-_64095120 | 5.54 |
ENSRNOT00000016837
|
Sestd1
|
SEC14 and spectrin domain containing 1 |
| chr6_+_44225233 | 5.49 |
ENSRNOT00000066593
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr20_+_27366213 | 5.43 |
ENSRNOT00000000302
ENSRNOT00000057939 |
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr3_-_52447622 | 5.31 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr5_+_58995249 | 5.07 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr1_+_99505677 | 5.02 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chrX_+_9956370 | 4.74 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr5_+_22392732 | 4.69 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr20_+_3830164 | 4.66 |
ENSRNOT00000045533
ENSRNOT00000084117 |
Col11a2
|
collagen type XI alpha 2 chain |
| chr16_+_48513432 | 4.64 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chrX_-_32095867 | 4.62 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr8_+_122076759 | 4.50 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr9_-_30251388 | 4.50 |
ENSRNOT00000035033
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr8_-_39460844 | 4.50 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_+_87224677 | 4.48 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_+_69841998 | 4.36 |
ENSRNOT00000072231
|
Zfp773-ps1
|
zinc finger protein 773, pseudogene 1 |
| chr11_+_53081025 | 4.35 |
ENSRNOT00000002700
|
Bbx
|
BBX, HMG-box containing |
| chr1_-_23556241 | 4.29 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr2_-_185852759 | 4.26 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr2_-_139528162 | 4.17 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr7_-_107203897 | 3.99 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr3_-_21904133 | 3.96 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr3_-_160561741 | 3.68 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr7_+_23854846 | 3.58 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr18_+_30880020 | 3.27 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr2_+_196462554 | 3.22 |
ENSRNOT00000056380
|
Fam63a
|
family with sequence similarity 63, member A |
| chr17_+_34704616 | 3.21 |
ENSRNOT00000090706
ENSRNOT00000083674 ENSRNOT00000077110 |
Exoc2
|
exocyst complex component 2 |
| chr8_-_33121002 | 3.12 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr6_+_106052212 | 3.10 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr8_+_71591360 | 3.04 |
ENSRNOT00000090096
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr8_-_7426611 | 3.02 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr10_-_71441389 | 2.97 |
ENSRNOT00000003699
|
Tada2a
|
transcriptional adaptor 2A |
| chr10_+_90230711 | 2.90 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_-_43638161 | 2.88 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr8_-_87419564 | 2.84 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr4_+_157524423 | 2.75 |
ENSRNOT00000036654
|
Zfp384
|
zinc finger protein 384 |
| chr4_+_29535852 | 2.69 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr17_-_2705123 | 2.60 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr12_+_25430464 | 2.58 |
ENSRNOT00000002020
|
Gtf2i
|
general transcription factor II I |
| chr8_-_69508024 | 2.58 |
ENSRNOT00000083564
|
AABR07070416.3
|
|
| chr11_-_61287914 | 2.40 |
ENSRNOT00000086286
|
Spice1
|
spindle and centriole associated protein 1 |
| chr2_+_226563050 | 2.38 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chrM_+_9870 | 2.29 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr1_-_212633304 | 2.25 |
ENSRNOT00000025611
|
Olr286
|
olfactory receptor 286 |
| chrX_+_144994139 | 2.25 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr4_-_131694755 | 2.23 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr3_+_77047466 | 2.19 |
ENSRNOT00000089424
|
Olr648
|
olfactory receptor 648 |
| chr11_+_70056624 | 2.14 |
ENSRNOT00000002447
|
AC133403.1
|
|
| chr5_+_6373583 | 2.08 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr18_+_30820321 | 2.07 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr17_-_14627937 | 2.07 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr1_+_217345545 | 1.95 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_+_141767940 | 1.93 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr2_-_170301348 | 1.92 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr3_+_172550258 | 1.89 |
ENSRNOT00000075148
|
Tubb1
|
tubulin, beta 1 class VI |
| chr1_-_241046249 | 1.85 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr17_+_11683862 | 1.82 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr3_-_45169118 | 1.79 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chrX_-_16532089 | 1.75 |
ENSRNOT00000093156
|
Dgkk
|
diacylglycerol kinase kappa |
| chr3_+_148654668 | 1.72 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr18_+_59748444 | 1.70 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr2_+_112914375 | 1.67 |
ENSRNOT00000092737
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr11_+_46737892 | 1.54 |
ENSRNOT00000052182
|
AABR07033979.1
|
|
| chr10_+_76320041 | 1.49 |
ENSRNOT00000037693
|
Coil
|
coilin |
| chr1_+_227892956 | 1.49 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr20_+_42966140 | 1.49 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr3_-_141411170 | 1.43 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr20_+_1172867 | 1.34 |
ENSRNOT00000041189
|
Olr1708
|
olfactory receptor 1708 |
| chr1_+_201337416 | 1.30 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr8_+_5833359 | 1.27 |
ENSRNOT00000033306
|
Mmp20
|
matrix metallopeptidase 20 |
| chr3_+_31802999 | 1.26 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
| chrX_-_70835089 | 1.25 |
ENSRNOT00000076351
|
Tex11
|
testis expressed 11 |
| chr3_-_90751055 | 1.21 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr1_+_86429262 | 1.16 |
ENSRNOT00000045789
|
Vom1r3
|
vomeronasal 1 receptor 3 |
| chr16_+_24978527 | 1.14 |
ENSRNOT00000019363
|
Tma16
|
translation machinery associated 16 homolog |
| chr18_-_74059533 | 1.05 |
ENSRNOT00000038767
|
RGD1308601
|
similar to hypothetical protein |
| chr9_+_71247781 | 1.03 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr10_+_61685645 | 0.88 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr6_+_2216623 | 0.87 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr1_+_255579474 | 0.79 |
ENSRNOT00000024465
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr8_-_20173107 | 0.72 |
ENSRNOT00000045215
|
Olr1163
|
olfactory receptor 1163 |
| chr4_+_2593876 | 0.70 |
ENSRNOT00000092992
|
Ube3c
|
ubiquitin protein ligase E3C |
| chr10_+_91126689 | 0.55 |
ENSRNOT00000004046
|
Nmt1
|
N-myristoyltransferase 1 |
| chr7_+_9262982 | 0.54 |
ENSRNOT00000047647
|
Olr1063
|
olfactory receptor 1063 |
| chr3_-_72081079 | 0.50 |
ENSRNOT00000007914
|
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chrX_-_106607352 | 0.47 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr4_+_6827429 | 0.46 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr15_-_55209342 | 0.44 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr11_+_43309759 | 0.35 |
ENSRNOT00000040089
|
Olr1539
|
olfactory receptor 1539 |
| chr18_-_26656879 | 0.35 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr12_-_19167015 | 0.34 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr10_+_95770154 | 0.31 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr7_-_3707226 | 0.28 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr16_-_74496731 | 0.25 |
ENSRNOT00000015790
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr7_+_58570579 | 0.21 |
ENSRNOT00000081227
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr10_-_62184874 | 0.17 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr7_-_4183525 | 0.14 |
ENSRNOT00000047673
|
Olr889
|
olfactory receptor 889 |
| chr8_+_5790034 | 0.12 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr1_-_165606375 | 0.06 |
ENSRNOT00000024290
|
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr8_+_22648323 | 0.05 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr6_+_123034304 | 0.04 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr5_-_28164326 | 0.04 |
ENSRNOT00000088165
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr10_+_88227360 | 0.03 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa7_Hoxc8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 63.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 5.4 | 21.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.7 | 27.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 2.0 | 13.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 1.8 | 5.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 1.8 | 7.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 1.6 | 6.3 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.6 | 15.5 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 1.5 | 4.6 | GO:0015827 | angiotensin-mediated drinking behavior(GO:0003051) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 1.4 | 5.7 | GO:0021888 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 1.2 | 49.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 1.0 | 3.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.9 | 5.5 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.9 | 12.6 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 5.7 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.8 | 4.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.7 | 2.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.7 | 2.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.6 | 1.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.6 | 9.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.6 | 1.7 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.5 | 5.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 6.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.5 | 7.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 8.5 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.5 | 1.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 3.2 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.4 | 4.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 17.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 2.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 1.0 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.3 | 5.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.3 | 1.8 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 | 4.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.3 | 3.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 4.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.3 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 1.2 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.2 | 1.5 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 5.8 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.2 | 6.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 2.0 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.2 | 4.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 10.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 3.7 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 0.4 | GO:1903944 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 2.4 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 6.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 1.9 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 20.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.8 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 4.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 0.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 3.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 3.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 17.0 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 15.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 4.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 1.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 1.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 13.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 3.6 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.9 | GO:0007569 | cell aging(GO:0007569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 54.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 2.6 | 69.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 2.1 | 6.3 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.6 | 4.7 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 1.5 | 13.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.9 | 15.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.7 | 5.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.6 | 4.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.6 | 5.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 3.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.4 | 6.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 3.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.4 | 1.8 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 17.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 6.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 14.5 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.2 | 7.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 6.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 3.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 24.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 1.2 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 4.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 4.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 5.0 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 11.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.4 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 8.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 10.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 4.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 5.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 8.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 6.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.8 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 60.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 4.3 | 21.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.2 | 49.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 2.1 | 6.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 1.5 | 7.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 1.4 | 4.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 1.4 | 5.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.2 | 4.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.1 | 17.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.7 | 5.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.6 | 1.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.6 | 5.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.6 | 1.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 12.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.4 | 13.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.4 | 5.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 10.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 11.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.3 | 6.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 1.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 9.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.2 | 8.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 3.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 4.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.0 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.1 | 4.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 10.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 1.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 7.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 7.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 4.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 4.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 3.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 15.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 2.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 14.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 3.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 4.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 5.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.5 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 6.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 17.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 6.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 7.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 12.1 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 46.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 2.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 13.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 15.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 3.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 5.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 70.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 5.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 25.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.4 | 5.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 9.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 1.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 5.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 5.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 6.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 6.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 7.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |