Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
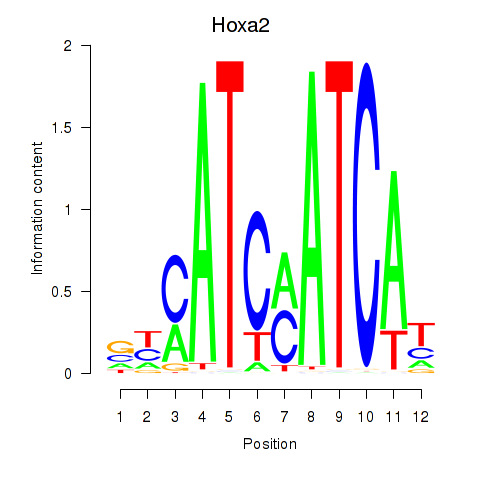
Results for Hoxa2
Z-value: 1.45
Transcription factors associated with Hoxa2
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Hoxa2 motif
Sorted Z-values of Hoxa2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_3293599 | 81.87 |
ENSRNOT00000081999
ENSRNOT00000047118 ENSRNOT00000020427 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr3_-_45169118 | 37.24 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr10_-_96131880 | 37.01 |
ENSRNOT00000004578
|
Cacng5
|
calcium voltage-gated channel auxiliary subunit gamma 5 |
| chr6_+_132242328 | 36.94 |
ENSRNOT00000081088
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr3_+_159368273 | 36.67 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr9_-_89193821 | 36.59 |
ENSRNOT00000090881
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr16_-_7007051 | 32.80 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr2_+_228544418 | 32.62 |
ENSRNOT00000013030
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr11_+_80358211 | 32.46 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr4_-_184096806 | 30.77 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr6_-_86223052 | 30.25 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr17_+_9746485 | 29.86 |
ENSRNOT00000072298
|
Pfn3
|
profilin 3 |
| chr17_-_43584152 | 29.36 |
ENSRNOT00000023241
|
Slc17a2
|
solute carrier family 17, member 2 |
| chr17_+_23661429 | 28.36 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr6_-_23291568 | 28.08 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr9_+_95501778 | 26.74 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr1_+_101161252 | 26.50 |
ENSRNOT00000028064
ENSRNOT00000064184 |
Slc17a7
|
solute carrier family 17 member 7 |
| chr1_+_150310319 | 26.46 |
ENSRNOT00000042081
|
Olr34
|
olfactory receptor 34 |
| chr1_+_219964429 | 26.43 |
ENSRNOT00000088288
|
Sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr17_+_88215834 | 26.37 |
ENSRNOT00000034098
|
Gpr158
|
G protein-coupled receptor 158 |
| chr16_+_34795971 | 26.35 |
ENSRNOT00000043510
|
LOC100912321
|
myeloid-associated differentiation marker-like |
| chr2_+_147496229 | 25.88 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr7_+_20262680 | 25.39 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr4_+_22859622 | 24.78 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr6_+_113898420 | 24.38 |
ENSRNOT00000064872
|
Nrxn3
|
neurexin 3 |
| chrX_-_142248369 | 24.16 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr9_-_89195273 | 24.03 |
ENSRNOT00000022281
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr1_+_229063714 | 24.03 |
ENSRNOT00000087526
|
Glyat
|
glycine-N-acyltransferase |
| chr1_-_67094567 | 23.17 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr14_+_22517774 | 22.38 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr16_-_75156321 | 22.33 |
ENSRNOT00000058063
|
Defb11
|
defensin beta 11 |
| chr7_-_136853957 | 22.31 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr6_+_26797126 | 21.83 |
ENSRNOT00000010586
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr10_-_8498422 | 21.78 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr9_-_4879755 | 21.72 |
ENSRNOT00000047615
|
RGD1559960
|
similar to Sulfotransferase K1 (rSULT1C2) |
| chrX_-_1848904 | 21.71 |
ENSRNOT00000010984
|
Rgn
|
regucalcin |
| chr9_+_4094995 | 21.67 |
ENSRNOT00000089450
|
LOC100910057
|
sulfotransferase 1C2-like |
| chr13_+_24823488 | 21.38 |
ENSRNOT00000019907
|
Cdh20
|
cadherin 20 |
| chrX_-_38468360 | 20.76 |
ENSRNOT00000045285
|
Map7d2
|
MAP7 domain containing 2 |
| chr8_-_8524643 | 20.50 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr7_-_100382897 | 20.37 |
ENSRNOT00000006510
|
LOC500876
|
LOC500876 |
| chr19_+_15294248 | 20.31 |
ENSRNOT00000024622
|
Ces1f
|
carboxylesterase 1F |
| chr19_+_6046665 | 20.06 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr6_+_112203679 | 19.98 |
ENSRNOT00000031205
|
Nrxn3
|
neurexin 3 |
| chr4_-_155275161 | 19.94 |
ENSRNOT00000032690
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr2_-_183210799 | 19.83 |
ENSRNOT00000085382
|
Trim2
|
tripartite motif-containing 2 |
| chr1_-_227392054 | 19.70 |
ENSRNOT00000054806
|
Ms4a13-ps1
|
membrane-spanning 4-domains, subfamily A, member 13, pseudogene 1 |
| chr20_+_20236151 | 19.68 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chrX_+_82143789 | 19.52 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr8_-_80631873 | 18.60 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr19_+_25983169 | 18.50 |
ENSRNOT00000004404
|
Syce2
|
synaptonemal complex central element protein 2 |
| chr7_-_106753592 | 18.47 |
ENSRNOT00000006930
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr13_+_77485113 | 18.44 |
ENSRNOT00000080254
|
Tnr
|
tenascin R |
| chr20_-_54517709 | 18.25 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr5_-_166116516 | 18.17 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr1_+_1702696 | 17.93 |
ENSRNOT00000019181
|
Lrp11
|
LDL receptor related protein 11 |
| chr16_+_56247659 | 17.93 |
ENSRNOT00000017452
|
Tusc3
|
tumor suppressor candidate 3 |
| chr7_+_123482255 | 17.90 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr7_+_34402738 | 17.77 |
ENSRNOT00000030985
|
Ccdc38
|
coiled-coil domain containing 38 |
| chr8_+_41657566 | 17.62 |
ENSRNOT00000042860
|
Rup2
|
urinary protein 2 |
| chr6_+_64808238 | 17.62 |
ENSRNOT00000093195
|
Nrcam
|
neuronal cell adhesion molecule |
| chr5_+_122390522 | 17.57 |
ENSRNOT00000064567
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr16_-_74330911 | 17.55 |
ENSRNOT00000084330
|
Slc20a2
|
solute carrier family 20 member 2 |
| chr1_-_67134827 | 17.51 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr3_+_69549673 | 17.35 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr2_-_173563273 | 17.24 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr4_+_96831880 | 17.19 |
ENSRNOT00000068400
|
RSA-14-44
|
RSA-14-44 protein |
| chr7_+_20462081 | 16.75 |
ENSRNOT00000088383
|
AABR07056131.1
|
|
| chr10_+_1920529 | 16.65 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr4_+_138269142 | 16.62 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chr2_-_210738378 | 16.40 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr9_-_4327679 | 16.25 |
ENSRNOT00000073468
|
LOC100910235
|
sulfotransferase 1C1-like |
| chr15_-_6587367 | 16.11 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr9_+_8399632 | 16.10 |
ENSRNOT00000092165
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chr6_+_97168453 | 15.83 |
ENSRNOT00000085790
|
Syt16
|
synaptotagmin 16 |
| chr4_+_30313102 | 15.81 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr9_+_118849302 | 15.79 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr6_+_10483308 | 15.76 |
ENSRNOT00000074516
|
Tmem247
|
transmembrane protein 247 |
| chr12_+_7502925 | 15.75 |
ENSRNOT00000072801
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr5_-_146446227 | 15.60 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr17_-_71897972 | 15.58 |
ENSRNOT00000065942
|
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr3_-_158328881 | 15.52 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr11_-_81735592 | 15.45 |
ENSRNOT00000078203
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr19_-_59991590 | 15.40 |
ENSRNOT00000091620
|
AABR07072668.1
|
|
| chr18_+_61563053 | 15.39 |
ENSRNOT00000022845
|
Grp
|
gastrin releasing peptide |
| chr5_+_124476168 | 15.27 |
ENSRNOT00000077754
|
RGD1564074
|
similar to novel protein |
| chr4_-_117589464 | 15.20 |
ENSRNOT00000021167
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chrX_+_105134498 | 15.16 |
ENSRNOT00000002058
|
Tmem35
|
transmembrane protein 35 |
| chr19_+_24545318 | 15.04 |
ENSRNOT00000005071
|
Clgn
|
calmegin |
| chr5_+_10178302 | 14.92 |
ENSRNOT00000009679
|
Sntg1
|
syntrophin, gamma 1 |
| chr9_+_73378057 | 14.92 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_+_40038336 | 14.70 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr2_+_114413410 | 14.69 |
ENSRNOT00000015866
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr7_-_124929025 | 14.69 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr14_+_22724399 | 14.57 |
ENSRNOT00000002724
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr1_+_125367280 | 14.45 |
ENSRNOT00000022049
|
Apba2
|
amyloid beta precursor protein binding family A member 2 |
| chr9_+_4817854 | 14.42 |
ENSRNOT00000040879
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr1_-_215838209 | 14.12 |
ENSRNOT00000050760
|
Igf2
|
insulin-like growth factor 2 |
| chr2_+_78247448 | 14.09 |
ENSRNOT00000089805
|
LOC103689968
|
protein FAM134B |
| chr4_+_64088900 | 14.08 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr4_+_122217505 | 14.02 |
ENSRNOT00000030426
|
LOC685964
|
hypothetical protein LOC685964 |
| chr1_-_224533219 | 13.96 |
ENSRNOT00000051289
|
Ust5r
|
integral membrane transport protein UST5r |
| chr12_-_52124779 | 13.89 |
ENSRNOT00000088839
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr9_+_73319710 | 13.87 |
ENSRNOT00000092485
|
Map2
|
microtubule-associated protein 2 |
| chr7_-_130827152 | 13.85 |
ENSRNOT00000019406
|
Syt10
|
synaptotagmin 10 |
| chr6_+_64297888 | 13.81 |
ENSRNOT00000050222
ENSRNOT00000083088 ENSRNOT00000093147 |
Nrcam
|
neuronal cell adhesion molecule |
| chrX_-_15707436 | 13.74 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr2_+_237679792 | 13.73 |
ENSRNOT00000064628
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr9_-_91683468 | 13.67 |
ENSRNOT00000041550
|
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr14_-_46054022 | 13.52 |
ENSRNOT00000002982
|
LOC498368
|
similar to RIKEN cDNA 0610040J01 |
| chr1_-_260992291 | 13.42 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr3_+_148790979 | 13.37 |
ENSRNOT00000085757
|
Kif3b
|
kinesin family member 3B |
| chr17_+_43458553 | 13.36 |
ENSRNOT00000088939
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr1_+_83714347 | 13.28 |
ENSRNOT00000085245
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr18_-_51651267 | 13.19 |
ENSRNOT00000020325
|
Aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr9_+_8349033 | 13.06 |
ENSRNOT00000073775
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chrX_-_115175299 | 13.02 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr17_-_48562838 | 13.00 |
ENSRNOT00000017102
ENSRNOT00000084702 |
Amph
|
amphiphysin |
| chr11_+_20474483 | 12.99 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr4_+_52129556 | 12.98 |
ENSRNOT00000009435
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chr9_+_4731914 | 12.97 |
ENSRNOT00000061893
|
AABR07066201.1
|
|
| chr5_+_124021366 | 12.97 |
ENSRNOT00000009977
|
Dab1
|
DAB1, reelin adaptor protein |
| chr13_-_42263024 | 12.65 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr19_-_62150423 | 12.63 |
ENSRNOT00000073524
ENSRNOT00000091209 |
Ccdc7
|
coiled-coil domain containing 7 |
| chr3_-_170040953 | 12.51 |
ENSRNOT00000005866
|
Cbln4
|
cerebellin 4 precursor |
| chr13_+_71331052 | 12.41 |
ENSRNOT00000075480
|
Glul
|
glutamate-ammonia ligase |
| chr18_-_15089988 | 12.37 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr9_-_4945352 | 12.30 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr10_+_88356615 | 12.24 |
ENSRNOT00000022687
|
Klhl10
|
kelch-like family member 10 |
| chr9_-_32868371 | 12.15 |
ENSRNOT00000038369
|
LOC689725
|
similar to chromosome 9 open reading frame 79 |
| chr3_-_64543100 | 12.04 |
ENSRNOT00000025803
|
Zfp385b
|
zinc finger protein 385B |
| chr5_-_134008255 | 12.04 |
ENSRNOT00000012448
|
Cyp4a8
|
cytochrome P450, family 4, subfamily a, polypeptide 8 |
| chr19_-_50220455 | 11.98 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr8_-_40883880 | 11.90 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr20_-_45024315 | 11.88 |
ENSRNOT00000066856
|
RGD1304770
|
similar to Na+ dependent glucose transporter 1 |
| chr8_+_117906014 | 11.81 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr10_+_103992309 | 11.75 |
ENSRNOT00000065292
|
Trim80
|
tripartite motif protein 80 |
| chr13_-_51076852 | 11.72 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr4_-_159192526 | 11.65 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr11_+_30363280 | 11.59 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr7_-_141185710 | 11.55 |
ENSRNOT00000085033
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr6_-_26445787 | 11.54 |
ENSRNOT00000084242
|
LOC103692570
|
dihydropyrimidinase-related protein 5-like |
| chr1_+_189364288 | 11.53 |
ENSRNOT00000080338
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr11_+_60102121 | 11.50 |
ENSRNOT00000045521
|
Tmprss7
|
transmembrane protease, serine 7 |
| chr12_+_30165694 | 11.37 |
ENSRNOT00000001211
|
Asl
|
argininosuccinate lyase |
| chr19_-_43841795 | 11.36 |
ENSRNOT00000079539
|
Ldhd
|
lactate dehydrogenase D |
| chr2_-_181531978 | 11.29 |
ENSRNOT00000072029
|
Npy2r
|
neuropeptide Y receptor Y2 |
| chr3_+_65672058 | 11.28 |
ENSRNOT00000057901
|
RGD1564319
|
similar to TF-1 apoptosis related protein 19 |
| chr1_+_282568287 | 11.20 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr17_-_57526461 | 11.20 |
ENSRNOT00000072723
|
LOC103690053
|
ankyrin repeat domain-containing protein 26-like |
| chr1_-_224389389 | 11.18 |
ENSRNOT00000077408
ENSRNOT00000050010 |
UST4r
|
integral membrane transport protein UST4r |
| chr9_+_12475006 | 11.11 |
ENSRNOT00000079703
|
LOC100912293
|
uncharacterized LOC100912293 |
| chr7_-_139063752 | 11.09 |
ENSRNOT00000072309
|
LOC102551901
|
protein lifeguard 2-like |
| chr17_+_42133076 | 11.05 |
ENSRNOT00000031384
|
Aldh5a1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr9_+_10760113 | 11.03 |
ENSRNOT00000073054
|
Arrdc5
|
arrestin domain containing 5 |
| chr9_+_95309759 | 10.86 |
ENSRNOT00000051438
|
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr13_-_93307199 | 10.83 |
ENSRNOT00000065041
|
Rgs7
|
regulator of G-protein signaling 7 |
| chr19_+_26106838 | 10.78 |
ENSRNOT00000035987
|
Hook2
|
hook microtubule-tethering protein 2 |
| chr6_-_133716847 | 10.75 |
ENSRNOT00000072399
|
Rtl1
|
retrotransposon-like 1 |
| chr10_-_107539465 | 10.72 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr2_-_235177275 | 10.71 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr1_-_71710374 | 10.68 |
ENSRNOT00000078556
ENSRNOT00000046152 |
Nlrp4
|
NLR family, pyrin domain containing 4 |
| chr4_-_31730386 | 10.68 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr1_-_148119857 | 10.65 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chrX_+_62282212 | 10.63 |
ENSRNOT00000039568
|
AABR07038837.1
|
|
| chr2_-_188645196 | 10.62 |
ENSRNOT00000083793
|
Efna3
|
ephrin A3 |
| chr13_-_37287458 | 10.59 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr3_+_117421604 | 10.58 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr3_-_120373500 | 10.57 |
ENSRNOT00000067727
|
Nphp1
|
nephrocystin 1 |
| chr2_+_68820615 | 10.55 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr4_-_157294047 | 10.51 |
ENSRNOT00000005601
|
Eno2
|
enolase 2 |
| chr15_+_108608204 | 10.46 |
ENSRNOT00000018918
|
Clybl
|
citrate lyase beta like |
| chr7_+_119647375 | 10.38 |
ENSRNOT00000046563
|
Kctd17
|
potassium channel tetramerization domain containing 17 |
| chr9_+_35289443 | 10.37 |
ENSRNOT00000037765
ENSRNOT00000060503 |
Spata31e1
|
SPATA31 subfamily E, member 1 |
| chr10_+_34414854 | 10.25 |
ENSRNOT00000074980
|
LOC684471
|
similar to olfactory receptor 1394 |
| chr2_-_172459165 | 10.22 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr9_+_20251521 | 10.05 |
ENSRNOT00000005535
|
LOC100911625
|
gamma-enolase-like |
| chr14_+_22072024 | 9.94 |
ENSRNOT00000002680
|
ste2
|
estrogen sulfotransferase |
| chr1_+_83163079 | 9.92 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chr15_+_17775692 | 9.89 |
ENSRNOT00000061169
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr7_-_93502571 | 9.83 |
ENSRNOT00000077033
ENSRNOT00000076080 |
Samd12
|
sterile alpha motif domain containing 12 |
| chr19_+_52258947 | 9.74 |
ENSRNOT00000021072
|
Adad2
|
adenosine deaminase domain containing 2 |
| chr3_+_75906945 | 9.73 |
ENSRNOT00000047110
|
Olr586
|
olfactory receptor 586 |
| chr1_-_189199376 | 9.68 |
ENSRNOT00000021027
|
Umod
|
uromodulin |
| chr8_+_53678994 | 9.67 |
ENSRNOT00000083419
|
Drd2
|
dopamine receptor D2 |
| chr1_+_98425903 | 9.67 |
ENSRNOT00000024170
|
LOC690483
|
hypothetical protein LOC690483 |
| chr2_+_57276919 | 9.65 |
ENSRNOT00000063899
|
RGD1310081
|
similar to hypothetical protein FLJ13231 |
| chr4_+_149908375 | 9.64 |
ENSRNOT00000019504
|
LOC100909657
|
uncharacterized LOC100909657 |
| chr5_-_77749613 | 9.58 |
ENSRNOT00000075988
|
Mup5
|
major urinary protein 5 |
| chrX_-_42329232 | 9.53 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr16_+_48120864 | 9.42 |
ENSRNOT00000068562
|
Stox2
|
storkhead box 2 |
| chr17_+_56935451 | 9.39 |
ENSRNOT00000058966
|
RGD1564129
|
similar to hypothetical protein 4930474N05 |
| chr3_+_70327193 | 9.38 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr17_+_10586043 | 9.37 |
ENSRNOT00000023125
|
LOC306766
|
hypothetical LOC306766 |
| chr7_-_107223047 | 9.33 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr3_-_2444281 | 9.27 |
ENSRNOT00000013863
|
Tubb4b
|
tubulin, beta 4B class IVb |
| chr3_+_63379031 | 9.22 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chrX_-_107542510 | 9.21 |
ENSRNOT00000074140
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chrX_+_71199491 | 9.21 |
ENSRNOT00000076168
ENSRNOT00000005077 ENSRNOT00000005102 |
Nlgn3
|
neuroligin 3 |
| chr11_-_81717521 | 9.15 |
ENSRNOT00000058422
|
Ahsg
|
alpha-2-HS-glycoprotein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.7 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 6.6 | 26.5 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 5.9 | 23.8 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 5.7 | 17.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 5.2 | 21.0 | GO:0048252 | lauric acid metabolic process(GO:0048252) |
| 4.9 | 19.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 4.6 | 4.6 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 4.6 | 13.7 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 4.4 | 17.5 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) negative regulation of dopamine receptor signaling pathway(GO:0060160) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 4.3 | 13.0 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 3.9 | 11.7 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 3.9 | 11.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 3.7 | 11.2 | GO:0016107 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 3.6 | 18.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 3.5 | 10.6 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 3.5 | 10.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 3.5 | 31.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 3.5 | 24.2 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 3.4 | 13.7 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 3.4 | 13.4 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 3.1 | 18.4 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.9 | 14.7 | GO:0015755 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 2.9 | 17.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 2.9 | 11.6 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 2.9 | 34.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 2.8 | 8.5 | GO:0021627 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 2.8 | 14.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 2.8 | 14.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 2.7 | 10.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.7 | 18.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 2.6 | 36.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 2.6 | 20.5 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 2.6 | 7.7 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 2.5 | 39.6 | GO:0051923 | sulfation(GO:0051923) |
| 2.4 | 9.7 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 2.4 | 43.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 2.3 | 11.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 2.3 | 20.6 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 2.2 | 6.7 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 2.2 | 6.6 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 2.1 | 6.4 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 2.1 | 6.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 2.1 | 6.4 | GO:0010034 | response to acetate(GO:0010034) |
| 2.1 | 6.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 2.0 | 12.2 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 2.0 | 14.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.0 | 10.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 2.0 | 46.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 2.0 | 15.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 2.0 | 7.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.9 | 13.3 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 1.9 | 7.6 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.9 | 17.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.9 | 61.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 1.9 | 5.6 | GO:0090370 | lysophospholipid transport(GO:0051977) negative regulation of cholesterol efflux(GO:0090370) |
| 1.9 | 7.5 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 1.9 | 22.3 | GO:0070050 | development of secondary female sexual characteristics(GO:0046543) neuron cellular homeostasis(GO:0070050) |
| 1.9 | 37.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 1.8 | 5.4 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.8 | 5.4 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 1.8 | 7.0 | GO:1904008 | response to monosodium glutamate(GO:1904008) |
| 1.7 | 10.3 | GO:0097688 | glutamate receptor clustering(GO:0097688) |
| 1.7 | 5.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 1.7 | 6.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 1.6 | 13.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 1.6 | 4.9 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 1.5 | 4.6 | GO:0060082 | eye blink reflex(GO:0060082) |
| 1.5 | 10.5 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 1.4 | 9.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.4 | 6.8 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.3 | 8.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 1.3 | 15.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 1.3 | 26.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.3 | 19.5 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 1.3 | 3.9 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 1.3 | 17.9 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 1.3 | 25.4 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 1.3 | 3.8 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 1.3 | 5.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 1.2 | 8.7 | GO:0022605 | oogenesis stage(GO:0022605) |
| 1.2 | 11.2 | GO:0015747 | urate transport(GO:0015747) |
| 1.2 | 3.7 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 1.2 | 15.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 1.2 | 4.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.2 | 6.0 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.2 | 9.7 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 1.2 | 15.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 1.2 | 9.5 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.2 | 4.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.2 | 10.5 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 1.2 | 3.5 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 1.2 | 18.5 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 1.1 | 4.5 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 1.1 | 32.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 1.1 | 30.5 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 1.1 | 32.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 1.1 | 24.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.1 | 7.7 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 1.1 | 6.6 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 1.1 | 7.7 | GO:0033572 | transferrin transport(GO:0033572) |
| 1.1 | 16.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 1.1 | 4.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.1 | 4.2 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 1.0 | 5.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.0 | 3.1 | GO:0048749 | compound eye development(GO:0048749) |
| 1.0 | 6.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 1.0 | 6.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.0 | 3.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.0 | 3.9 | GO:0031179 | peptide modification(GO:0031179) |
| 0.9 | 13.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.9 | 1.8 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.9 | 3.6 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.9 | 2.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.9 | 7.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.9 | 28.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.9 | 13.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.9 | 11.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.8 | 6.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.8 | 2.4 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) basophil activation(GO:0045575) |
| 0.8 | 17.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.8 | 3.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.8 | 2.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.8 | 3.8 | GO:0031296 | cobalamin transport(GO:0015889) B cell costimulation(GO:0031296) |
| 0.7 | 3.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.7 | 12.6 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.7 | 2.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 5.9 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.7 | 2.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.7 | 11.5 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.7 | 7.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.7 | 4.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.7 | 9.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.7 | 2.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.7 | 2.0 | GO:0006214 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 0.7 | 21.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.7 | 3.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.6 | 7.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.6 | 101.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.6 | 7.1 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) serine phosphorylation of STAT protein(GO:0042501) |
| 0.6 | 5.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.6 | 4.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.6 | 5.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.6 | 11.8 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.6 | 1.8 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.6 | 8.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.6 | 5.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.6 | 4.7 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.6 | 1.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.6 | 29.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 7.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 6.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 7.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.5 | 13.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.5 | 8.7 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.5 | 10.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.5 | 2.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.5 | 3.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.5 | 5.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.5 | 1.9 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.5 | 5.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 2.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.5 | 10.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.4 | 4.9 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.4 | 8.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.4 | 8.0 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.4 | 2.2 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.4 | 15.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.4 | 2.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 21.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.4 | 33.7 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.4 | 8.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.4 | 3.9 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 1.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.4 | 2.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 3.8 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.4 | 1.9 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.4 | 10.6 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.4 | 6.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.4 | 4.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.4 | 38.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 6.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 13.0 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 20.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.3 | 14.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.3 | 5.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.3 | 6.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 13.8 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.3 | 7.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 2.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 10.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.3 | 8.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 7.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 2.5 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.3 | 2.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 3.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.3 | 0.6 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.3 | 6.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.3 | 8.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 2.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.3 | 4.5 | GO:0007616 | long-term memory(GO:0007616) |
| 0.3 | 0.8 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.3 | 2.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 10.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 1.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 6.5 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.3 | 25.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.3 | 3.2 | GO:0048672 | bleb assembly(GO:0032060) positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 4.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 34.7 | GO:0007411 | axon guidance(GO:0007411) |
| 0.2 | 3.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 3.5 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 | 0.7 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 6.5 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 5.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 9.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.2 | 5.2 | GO:0071450 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.2 | 6.9 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 22.4 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.2 | 0.6 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 0.2 | 2.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 3.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 7.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 6.2 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.2 | 8.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.2 | 4.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 5.6 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.2 | 235.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 0.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 8.1 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.2 | 6.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 11.5 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.2 | 8.0 | GO:0032106 | positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.2 | 12.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 4.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 1.0 | GO:1903541 | regulation of exosomal secretion(GO:1903541) |
| 0.2 | 18.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.2 | 1.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 29.9 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 0.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 4.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 2.4 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 4.9 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 2.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.4 | GO:0046368 | GDP-L-fucose biosynthetic process(GO:0042350) GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 9.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 8.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 5.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 19.4 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.1 | 2.7 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 4.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 2.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 4.1 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 | 0.4 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 | 0.5 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 4.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.2 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990441) |
| 0.1 | 0.6 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 1.5 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.8 | GO:0042993 | positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.0 | 5.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 5.9 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 2.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.2 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 3.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 3.2 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 3.5 | 10.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 3.4 | 20.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 3.4 | 10.3 | GO:0098982 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 3.3 | 26.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 3.2 | 28.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 3.0 | 18.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 2.8 | 14.2 | GO:0070695 | FHF complex(GO:0070695) |
| 2.7 | 69.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 2.7 | 13.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 2.7 | 26.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 2.6 | 23.1 | GO:0008091 | spectrin(GO:0008091) |
| 2.3 | 14.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 2.3 | 7.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 2.0 | 18.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 2.0 | 15.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 2.0 | 110.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 1.9 | 13.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.8 | 30.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.6 | 6.5 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.6 | 21.1 | GO:0000801 | central element(GO:0000801) |
| 1.6 | 6.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 1.5 | 15.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.4 | 7.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.4 | 21.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.2 | 37.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.2 | 5.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.2 | 17.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 1.0 | 11.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 1.0 | 8.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.9 | 11.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.8 | 11.6 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.7 | 12.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.7 | 3.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.7 | 11.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 4.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.7 | 6.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.7 | 7.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.6 | 6.2 | GO:0070187 | telosome(GO:0070187) |
| 0.6 | 34.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.6 | 7.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.6 | 10.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.5 | 10.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.5 | 7.9 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.5 | 23.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.5 | 20.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.5 | 9.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.5 | 32.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.5 | 8.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 10.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 9.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 3.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.4 | 9.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 8.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.4 | 1.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 3.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 3.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 5.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 2.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 2.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 6.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 3.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 5.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 15.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.3 | 21.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 1.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 28.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 1.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 7.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 4.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.3 | 3.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 6.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 15.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 41.3 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 11.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 7.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 14.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 8.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 25.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 15.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 24.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 62.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.2 | 9.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 2.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 31.2 | GO:0097223 | sperm part(GO:0097223) |
| 0.2 | 10.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 9.3 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 1.4 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.2 | 23.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 18.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 7.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.2 | 2.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 15.1 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 55.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 7.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 32.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 3.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 3.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 93.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 1.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 5.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 9.2 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 6.7 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 258.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.9 | 50.7 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 8.8 | 44.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 6.2 | 24.6 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 5.4 | 37.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 4.7 | 14.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 4.6 | 46.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 4.0 | 24.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.9 | 11.6 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 3.8 | 15.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 3.7 | 11.2 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 3.5 | 10.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 3.5 | 31.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 3.4 | 20.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 3.4 | 10.3 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 3.0 | 18.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 3.0 | 8.9 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 2.8 | 8.3 | GO:0031765 | galanin receptor binding(GO:0031763) type 2 galanin receptor binding(GO:0031765) type 3 galanin receptor binding(GO:0031766) |
| 2.7 | 18.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 2.6 | 18.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 2.5 | 7.6 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 2.5 | 7.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 2.3 | 11.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 2.3 | 11.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 2.3 | 11.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 2.2 | 8.9 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 2.2 | 6.6 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 2.1 | 6.4 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 2.1 | 10.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 2.1 | 10.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 2.0 | 6.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 2.0 | 7.9 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.0 | 7.9 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 1.9 | 36.7 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.9 | 13.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.9 | 17.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.8 | 5.4 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.8 | 5.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 1.7 | 15.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 1.7 | 39.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.6 | 4.9 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 1.6 | 13.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 1.6 | 9.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.6 | 6.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 1.6 | 45.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.5 | 14.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 1.5 | 39.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.4 | 11.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 1.4 | 11.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.4 | 5.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.3 | 14.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 1.3 | 5.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.3 | 5.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.2 | 15.8 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 1.2 | 17.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 1.2 | 15.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.2 | 4.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.2 | 3.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.1 | 4.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 1.1 | 4.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.1 | 4.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.1 | 9.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 1.1 | 21.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.1 | 8.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 1.1 | 5.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.1 | 10.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 1.1 | 6.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.0 | 28.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.0 | 10.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.0 | 42.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 1.0 | 4.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.0 | 36.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 1.0 | 8.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.0 | 10.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.0 | 15.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.0 | 61.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 1.0 | 3.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.9 | 2.8 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.9 | 7.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.9 | 3.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.9 | 10.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.8 | 10.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.8 | 2.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.8 | 13.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.8 | 5.7 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.8 | 13.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.8 | 9.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.8 | 3.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.8 | 2.4 | GO:0099567 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 0.8 | 4.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.8 | 6.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.8 | 36.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.8 | 10.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.8 | 1.6 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.7 | 3.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.7 | 2.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.7 | 7.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.7 | 7.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.7 | 7.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.7 | 2.0 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.7 | 5.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.7 | 5.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.7 | 5.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.7 | 4.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.7 | 9.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.6 | 26.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.6 | 5.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.6 | 7.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.6 | 7.0 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.6 | 16.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.6 | 34.5 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.6 | 7.5 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.6 | 15.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.6 | 11.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 6.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.5 | 3.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 70.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.5 | 3.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 13.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.5 | 8.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.5 | 53.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.5 | 9.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.5 | 16.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 12.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 4.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.5 | 10.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 2.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 9.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.5 | 7.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 3.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 3.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.4 | 7.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.4 | 31.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.4 | 11.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.4 | 29.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.4 | 22.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 16.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 8.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.4 | 33.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.4 | 1.5 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.4 | 7.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 1.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 3.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.4 | 7.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 5.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 37.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.4 | 58.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.4 | 9.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 10.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 19.1 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.3 | 2.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 4.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.3 | 4.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 1.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.3 | 9.5 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.3 | 1.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.3 | 109.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.3 | 5.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.3 | 2.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 5.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 1.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) potassium ion antiporter activity(GO:0022821) |
| 0.2 | 6.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 3.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 16.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 1.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 0.6 | GO:0030283 | testosterone dehydrogenase [NAD(P)] activity(GO:0030283) |
| 0.2 | 4.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 22.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.2 | 2.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 4.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 2.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 4.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 2.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 2.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 5.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 126.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 4.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 30.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 4.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 14.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 7.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 28.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 2.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 7.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 12.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 5.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 12.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 9.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 6.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 0.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 4.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 5.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 9.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 7.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.3 | GO:0003921 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.1 | 6.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 6.1 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.1 | 2.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 2.3 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.8 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 6.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 3.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 47.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.7 | 26.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.6 | 33.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.6 | 3.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.5 | 6.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 20.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.4 | 4.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.4 | 23.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 25.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.4 | 9.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.4 | 11.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.4 | 9.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 13.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 8.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 11.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 9.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 4.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 10.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 36.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 2.6 | 92.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 2.5 | 7.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.3 | 20.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.2 | 26.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.0 | 40.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 1.0 | 20.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 1.0 | 17.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.9 | 11.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.9 | 13.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.8 | 4.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.8 | 19.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.8 | 14.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 11.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.7 | 22.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.7 | 5.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.7 | 33.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.7 | 42.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.7 | 14.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 34.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.6 | 10.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.5 | 7.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 10.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.5 | 7.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 13.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 9.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.5 | 6.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 9.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.4 | 5.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 15.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 6.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 10.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.4 | 9.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.4 | 6.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 5.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 11.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 7.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.3 | 8.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 6.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 8.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 8.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.3 | 4.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 2.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.3 | 4.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 7.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 2.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 1.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 13.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 25.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 3.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 11.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 5.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 2.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 10.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.2 | 7.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 5.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.2 | 0.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 4.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 2.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 13.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 4.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 5.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 7.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 1.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.4 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 5.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 4.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |