Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
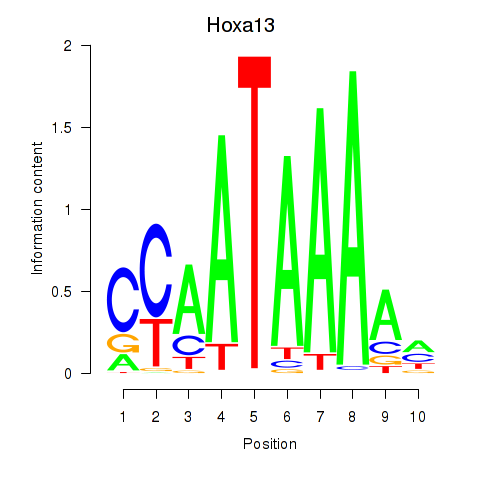
Results for Hoxa13
Z-value: 0.96
Transcription factors associated with Hoxa13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa13
|
ENSRNOG00000057061 | homeo box A13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa13 | rn6_v1_chr4_-_82229397_82229397 | 0.16 | 4.7e-03 | Click! |
Activity profile of Hoxa13 motif
Sorted Z-values of Hoxa13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_5773036 | 33.34 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr2_+_223029559 | 32.70 |
ENSRNOT00000045584
|
AABR07013111.1
|
|
| chr12_-_2438817 | 30.78 |
ENSRNOT00000037059
|
Ccl25
|
C-C motif chemokine ligand 25 |
| chr1_+_12915734 | 27.55 |
ENSRNOT00000089066
|
Txlnb
|
taxilin beta |
| chr1_-_16687817 | 26.82 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr17_-_43626538 | 26.12 |
ENSRNOT00000074292
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr3_-_60166013 | 25.78 |
ENSRNOT00000024922
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr14_-_71814523 | 25.61 |
ENSRNOT00000004094
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr4_-_163445302 | 24.03 |
ENSRNOT00000087106
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr4_+_78371121 | 23.46 |
ENSRNOT00000059157
|
Gimap1
|
GTPase, IMAP family member 1 |
| chr7_+_38742051 | 22.93 |
ENSRNOT00000006070
|
Dcn
|
decorin |
| chr2_+_93792601 | 22.31 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr17_+_47870611 | 21.79 |
ENSRNOT00000078555
|
AABR07027872.1
|
|
| chr1_+_8310577 | 20.67 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr1_+_219403970 | 20.05 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chrX_-_77675487 | 19.48 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chrX_-_10031167 | 19.07 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr11_-_45124423 | 18.43 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr4_+_70572942 | 18.38 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr5_+_90338795 | 17.26 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr2_+_183674522 | 17.11 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr4_+_44597123 | 17.01 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr2_-_88553086 | 16.99 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr12_-_40590361 | 16.87 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr4_-_109532234 | 16.54 |
ENSRNOT00000008665
|
Reg3g
|
regenerating family member 3 gamma |
| chr4_-_170860225 | 16.35 |
ENSRNOT00000007577
|
Mgp
|
matrix Gla protein |
| chr16_+_2537248 | 16.32 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr15_+_33606124 | 16.21 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr1_-_141471010 | 16.00 |
ENSRNOT00000082164
|
Plin1
|
perilipin 1 |
| chr9_-_52238564 | 15.95 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr13_-_61070599 | 15.79 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr18_+_17091310 | 15.59 |
ENSRNOT00000093285
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr2_-_208215187 | 15.50 |
ENSRNOT00000047836
|
Rap1a
|
RAP1A, member of RAS oncogene family |
| chr4_+_14151343 | 14.38 |
ENSRNOT00000061687
ENSRNOT00000076573 ENSRNOT00000077219 ENSRNOT00000008319 |
Cd36
|
CD36 molecule |
| chr10_+_57251253 | 14.36 |
ENSRNOT00000043171
|
LOC108348287
|
60S ribosomal protein L36a |
| chr2_-_257484587 | 14.10 |
ENSRNOT00000083344
|
Nexn
|
nexilin (F actin binding protein) |
| chr3_+_17107861 | 13.10 |
ENSRNOT00000043097
|
AABR07051563.1
|
|
| chr11_+_86421106 | 12.99 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr7_-_107391184 | 12.96 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr13_-_78521911 | 12.84 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr8_-_109560747 | 12.56 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr10_-_31493419 | 12.41 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr7_-_14418950 | 12.40 |
ENSRNOT00000066722
|
Rasal3
|
RAS protein activator like 3 |
| chr19_-_6293 | 12.39 |
ENSRNOT00000074240
|
LOC679149
|
similar to carboxylesterase 2 (intestine, liver) |
| chr6_+_132551394 | 12.26 |
ENSRNOT00000019583
|
Evl
|
Enah/Vasp-like |
| chr20_-_18060661 | 12.24 |
ENSRNOT00000070925
|
AABR07044711.1
|
|
| chr9_-_81400987 | 12.07 |
ENSRNOT00000035277
|
Tns1
|
tensin 1 |
| chr4_-_163445136 | 12.03 |
ENSRNOT00000080299
|
Klrc3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr2_-_117666683 | 11.85 |
ENSRNOT00000015479
|
AABR07009931.1
|
|
| chr11_+_85508300 | 11.84 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr17_+_32973695 | 11.78 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr4_+_155313671 | 11.71 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chr10_-_56558487 | 11.69 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr17_-_18590536 | 11.56 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_+_248178389 | 11.23 |
ENSRNOT00000037339
|
Gbp5
|
guanylate binding protein 5 |
| chr7_+_44146237 | 10.95 |
ENSRNOT00000058842
|
Rassf9
|
Ras association domain family member 9 |
| chr4_+_162493908 | 10.92 |
ENSRNOT00000072064
|
Clec2d
|
C-type lectin domain family 2, member D |
| chrX_+_51286737 | 10.86 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr13_-_92089980 | 10.70 |
ENSRNOT00000036040
|
Mnda
|
myeloid cell nuclear differentiation antigen |
| chr17_-_84247038 | 10.70 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr4_-_51199570 | 10.68 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr14_-_72122158 | 10.67 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_-_204087001 | 10.65 |
ENSRNOT00000051853
|
Cpxm2
|
carboxypeptidase X (M14 family), member 2 |
| chr14_+_83510640 | 10.56 |
ENSRNOT00000089928
ENSRNOT00000082242 ENSRNOT00000025378 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr9_-_9702306 | 10.46 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr17_-_65955606 | 10.40 |
ENSRNOT00000067949
ENSRNOT00000023601 |
Ryr2
|
ryanodine receptor 2 |
| chr12_-_13668515 | 10.34 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr13_-_51297621 | 10.23 |
ENSRNOT00000030926
|
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chr1_-_25839198 | 10.23 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr10_+_764421 | 10.21 |
ENSRNOT00000084608
ENSRNOT00000087567 |
Myh11
|
myosin heavy chain 11 |
| chr4_+_157726941 | 10.20 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr5_-_16799776 | 10.16 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr14_-_62595854 | 10.09 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr4_-_180234804 | 10.08 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chrM_+_3904 | 10.07 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr20_+_4363152 | 9.91 |
ENSRNOT00000000508
ENSRNOT00000084841 ENSRNOT00000072848 ENSRNOT00000077561 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr4_+_64088900 | 9.91 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr17_+_43616247 | 9.75 |
ENSRNOT00000072019
|
Hist1h3a
|
histone cluster 1, H3a |
| chr7_+_34533543 | 9.75 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr11_+_80255790 | 9.65 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr16_-_88347369 | 9.63 |
ENSRNOT00000019797
|
Pdha1l1
|
pyruvate dehydrogenase (lipoamide) alpha 1-like 1 |
| chr2_+_148262110 | 9.58 |
ENSRNOT00000046641
|
AABR07010705.1
|
|
| chr15_-_34693034 | 9.58 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr4_-_163214678 | 9.49 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr3_-_21061070 | 9.42 |
ENSRNOT00000044461
|
Olr422
|
olfactory receptor 422 |
| chr3_+_72385666 | 9.18 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr15_+_30570483 | 9.17 |
ENSRNOT00000071740
|
AABR07017745.1
|
|
| chr11_-_46369577 | 9.17 |
ENSRNOT00000049859
ENSRNOT00000047817 ENSRNOT00000002220 ENSRNOT00000002223 ENSRNOT00000063864 ENSRNOT00000070871 ENSRNOT00000072140 |
Abi3bp
|
ABI family member 3 binding protein |
| chr10_+_72272248 | 9.08 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr2_-_45518502 | 9.07 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr10_-_107386072 | 9.05 |
ENSRNOT00000004290
|
Timp2
|
TIMP metallopeptidase inhibitor 2 |
| chr4_-_163402561 | 8.95 |
ENSRNOT00000091890
|
Klrk1
|
killer cell lectin like receptor K1 |
| chr9_-_88534710 | 8.95 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr9_+_52023295 | 8.76 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr5_-_174921 | 8.75 |
ENSRNOT00000056239
|
Tipinl1
|
TIMELESS interacting protein like 1 |
| chr15_-_95514259 | 8.68 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr5_-_146795866 | 8.60 |
ENSRNOT00000065640
|
Tlr12
|
toll-like receptor 12 |
| chr3_+_53563194 | 8.54 |
ENSRNOT00000048300
|
Xirp2
|
xin actin-binding repeat containing 2 |
| chr2_+_236625357 | 8.53 |
ENSRNOT00000081248
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr14_+_83510278 | 8.51 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr10_-_70337532 | 8.49 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr4_+_14070553 | 8.46 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chr17_+_47241017 | 8.42 |
ENSRNOT00000092193
|
Gpr141
|
G protein-coupled receptor 141 |
| chr2_-_205212681 | 8.38 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr2_-_170301348 | 8.36 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr13_-_26769374 | 8.25 |
ENSRNOT00000003768
|
Bcl2
|
BCL2, apoptosis regulator |
| chr2_+_40554146 | 8.23 |
ENSRNOT00000015138
|
Pde4d
|
phosphodiesterase 4D |
| chr16_-_10941414 | 8.19 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr2_+_45182402 | 8.06 |
ENSRNOT00000060673
|
RGD1561161
|
similar to BC067074 protein |
| chr13_+_57243877 | 7.94 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr15_+_31417147 | 7.93 |
ENSRNOT00000092182
|
AABR07017824.2
|
|
| chr2_+_198359754 | 7.89 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr18_+_15738809 | 7.86 |
ENSRNOT00000075444
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr5_+_118418799 | 7.86 |
ENSRNOT00000012052
|
Alg6
|
ALG6, alpha-1,3-glucosyltransferase |
| chr7_-_143228060 | 7.82 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr3_+_149624712 | 7.78 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr4_-_170740274 | 7.75 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr2_-_187909394 | 7.71 |
ENSRNOT00000032355
|
Rab25
|
RAB25, member RAS oncogene family |
| chr5_-_150167077 | 7.67 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr15_-_48284548 | 7.66 |
ENSRNOT00000038336
|
Hmbox1
|
homeobox containing 1 |
| chr1_-_62558033 | 7.64 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr1_+_75428027 | 7.60 |
ENSRNOT00000058334
|
Pla2g4c
|
phospholipase A2, group IVC |
| chr5_-_60191941 | 7.58 |
ENSRNOT00000033373
|
Pax5
|
paired box 5 |
| chr4_-_82209933 | 7.52 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr2_+_38230757 | 7.51 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chrX_+_28539158 | 7.31 |
ENSRNOT00000073535
|
Tlr8
|
toll-like receptor 8 |
| chr14_-_34561696 | 7.28 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr16_-_74700815 | 7.27 |
ENSRNOT00000081297
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chr11_+_85532526 | 7.24 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr17_-_44758170 | 7.20 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr8_-_133128290 | 7.17 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr1_-_69835185 | 7.11 |
ENSRNOT00000020757
|
LOC108349745
|
spindle assembly abnormal protein 6 homolog |
| chr16_-_14360555 | 7.09 |
ENSRNOT00000065460
|
LOC290595
|
hypothetical gene supported by AF152002 |
| chrX_+_78042859 | 7.00 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr20_+_3364814 | 6.97 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr6_-_141321108 | 6.94 |
ENSRNOT00000040556
|
AABR07065789.3
|
|
| chr4_-_82295470 | 6.93 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr15_-_49773178 | 6.91 |
ENSRNOT00000090328
|
AABR07018272.1
|
|
| chr2_-_210116038 | 6.90 |
ENSRNOT00000074950
|
LOC684509
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr10_-_15211325 | 6.84 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr7_-_73130740 | 6.84 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr2_-_31753528 | 6.79 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr10_+_54155876 | 6.76 |
ENSRNOT00000072855
|
Gas7
|
growth arrest specific 7 |
| chr2_-_86475096 | 6.75 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr6_-_142585188 | 6.75 |
ENSRNOT00000067437
|
AABR07065815.1
|
|
| chr4_-_173640684 | 6.74 |
ENSRNOT00000010728
|
Rergl
|
RERG like |
| chrX_+_77119911 | 6.71 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chrM_-_14061 | 6.71 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr8_+_117906014 | 6.70 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr8_+_118066988 | 6.69 |
ENSRNOT00000056161
|
Map4
|
microtubule-associated protein 4 |
| chr1_+_199352344 | 6.58 |
ENSRNOT00000026466
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chrX_-_139916883 | 6.55 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr5_-_50068706 | 6.54 |
ENSRNOT00000084643
|
Orc3
|
origin recognition complex, subunit 3 |
| chr9_-_69878706 | 6.51 |
ENSRNOT00000035879
|
Ino80d
|
INO80 complex subunit D |
| chr9_+_20048121 | 6.50 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr3_+_110855000 | 6.47 |
ENSRNOT00000081613
|
Knl1
|
kinetochore scaffold 1 |
| chr1_+_199351628 | 6.43 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr1_-_129217058 | 6.42 |
ENSRNOT00000086439
|
Pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr10_-_37311625 | 6.42 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chrX_+_156429585 | 6.41 |
ENSRNOT00000083203
ENSRNOT00000077322 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr1_-_124803363 | 6.41 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr2_-_45077219 | 6.41 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr2_+_113984646 | 6.37 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr2_-_96509424 | 6.32 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr15_+_31456265 | 6.29 |
ENSRNOT00000075092
|
AABR07017825.1
|
|
| chr16_+_85331866 | 6.28 |
ENSRNOT00000019615
|
Lig4
|
DNA ligase 4 |
| chr8_-_49158971 | 6.26 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr3_+_161236898 | 6.24 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr1_+_42121636 | 6.23 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr2_-_231648122 | 6.22 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr3_-_7796385 | 6.20 |
ENSRNOT00000082489
|
Ntng2
|
netrin G2 |
| chr16_+_31734944 | 6.19 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr14_-_45165207 | 6.19 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr6_+_55812747 | 6.18 |
ENSRNOT00000008106
|
Sostdc1
|
sclerostin domain containing 1 |
| chr1_+_137860946 | 6.17 |
ENSRNOT00000031334
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr3_+_17889972 | 6.17 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr17_-_389967 | 6.12 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr15_+_31097291 | 6.11 |
ENSRNOT00000075456
|
AABR07017789.1
|
|
| chr12_-_38916237 | 6.03 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr8_+_82037977 | 6.02 |
ENSRNOT00000082288
|
Myo5a
|
myosin VA |
| chr6_-_86914883 | 6.01 |
ENSRNOT00000091202
|
Mis18bp1
|
MIS18 binding protein 1 |
| chr15_+_32324830 | 5.93 |
ENSRNOT00000071598
|
AABR07017895.1
|
|
| chr3_+_44025300 | 5.90 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr1_+_174862698 | 5.89 |
ENSRNOT00000013276
|
Swap70
|
SWAP switching B-cell complex 70 |
| chr3_+_151126591 | 5.88 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr7_-_18793289 | 5.88 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chr3_+_100373439 | 5.83 |
ENSRNOT00000085176
|
Kif18a
|
kinesin family member 18A |
| chr1_-_169534896 | 5.78 |
ENSRNOT00000082645
|
Trim30c
|
tripartite motif-containing 30C |
| chr2_+_113984824 | 5.77 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr9_+_82741920 | 5.72 |
ENSRNOT00000027337
|
Slc4a3
|
solute carrier family 4 member 3 |
| chr6_-_54020406 | 5.72 |
ENSRNOT00000086038
|
Hdac9
|
histone deacetylase 9 |
| chr1_-_60407295 | 5.71 |
ENSRNOT00000078350
|
Vom1r12
|
vomeronasal 1 receptor 12 |
| chr18_-_74478341 | 5.69 |
ENSRNOT00000074572
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr3_-_16750564 | 5.66 |
ENSRNOT00000084111
|
AABR07051548.1
|
|
| chrX_-_157200981 | 5.66 |
ENSRNOT00000087364
|
AABR07071935.1
|
|
| chr12_-_51877624 | 5.66 |
ENSRNOT00000056800
|
Chek2
|
checkpoint kinase 2 |
| chr9_-_45206427 | 5.66 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr12_-_22472044 | 5.65 |
ENSRNOT00000073356
|
Ufsp1
|
UFM1-specific peptidase 1 |
| chr3_-_24601063 | 5.64 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr1_-_89483988 | 5.62 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 26.8 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 8.3 | 33.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 6.2 | 30.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 5.9 | 35.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 5.7 | 17.0 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) regulation of inward rectifier potassium channel activity(GO:1901979) positive regulation of gap junction assembly(GO:1903598) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 4.2 | 12.6 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 4.0 | 15.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 3.8 | 22.8 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 3.6 | 10.9 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 3.6 | 10.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 3.5 | 10.4 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 3.3 | 9.9 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 3.3 | 6.5 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 3.2 | 9.7 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 3.1 | 9.4 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 3.1 | 15.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 3.1 | 9.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 3.0 | 8.9 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 2.9 | 22.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 2.8 | 11.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 2.8 | 8.3 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 2.7 | 8.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 2.6 | 5.1 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 2.4 | 7.3 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 2.4 | 9.7 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 2.3 | 13.7 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 2.2 | 8.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.2 | 6.5 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.1 | 6.3 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 2.1 | 6.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 2.0 | 24.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 2.0 | 9.9 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 2.0 | 5.9 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 1.9 | 5.6 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 1.8 | 5.4 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 1.8 | 5.3 | GO:0097213 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 1.7 | 6.7 | GO:0051542 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.6 | 4.9 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 1.6 | 7.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 1.6 | 6.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 1.6 | 18.8 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 1.6 | 4.7 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 1.6 | 6.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 1.6 | 7.8 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 1.6 | 6.2 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 1.5 | 6.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.5 | 6.0 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.5 | 4.5 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 1.5 | 4.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.5 | 8.7 | GO:0048478 | replication fork protection(GO:0048478) |
| 1.5 | 7.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 1.4 | 4.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 1.4 | 27.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 1.4 | 5.7 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 1.4 | 4.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.4 | 4.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 1.4 | 5.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.3 | 9.1 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 1.3 | 27.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 1.2 | 12.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 1.2 | 7.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.2 | 8.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.2 | 3.5 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 1.2 | 4.7 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 1.2 | 3.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.1 | 30.9 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.1 | 3.4 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 1.1 | 1.1 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 1.1 | 4.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 1.1 | 3.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 1.1 | 12.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.0 | 15.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.0 | 6.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.0 | 3.0 | GO:2000660 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.0 | 10.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 1.0 | 6.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 1.0 | 3.9 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.9 | 10.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.9 | 5.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.9 | 5.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.9 | 13.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.9 | 3.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.9 | 2.7 | GO:0010643 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 0.9 | 2.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.9 | 12.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.8 | 3.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.8 | 5.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.8 | 2.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.8 | 14.4 | GO:0009750 | response to fructose(GO:0009750) |
| 0.8 | 3.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.8 | 11.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.8 | 3.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.8 | 2.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.8 | 3.8 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.8 | 5.3 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.8 | 3.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.7 | 3.7 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.7 | 3.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.7 | 7.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.7 | 6.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.7 | 5.6 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.7 | 7.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.7 | 4.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.7 | 4.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.7 | 4.7 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.7 | 25.6 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.6 | 3.2 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.6 | 3.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.6 | 9.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.6 | 2.5 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.6 | 3.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.6 | 3.7 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.6 | 3.7 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.6 | 3.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.6 | 3.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.6 | 5.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.6 | 1.7 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.6 | 3.5 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.6 | 2.9 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.6 | 4.6 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.6 | 2.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 5.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.6 | 10.5 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.5 | 1.6 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.5 | 2.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.5 | 7.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.5 | 5.4 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.5 | 0.5 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.5 | 4.8 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.5 | 3.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 | 3.6 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.5 | 10.7 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.5 | 7.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.5 | 11.7 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.5 | 10.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.5 | 10.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.5 | 1.5 | GO:0061341 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.5 | 6.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.5 | 3.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 1.9 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.5 | 6.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.5 | 4.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.5 | 10.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.5 | 4.1 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.5 | 8.1 | GO:0007567 | parturition(GO:0007567) |
| 0.4 | 1.3 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.4 | 3.6 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.4 | 6.9 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 7.7 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.4 | 17.0 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.4 | 2.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 | 6.7 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.4 | 1.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.4 | 1.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.4 | 8.6 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.4 | 2.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 2.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.4 | 2.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 7.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.4 | 1.9 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.4 | 2.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.4 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 1.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 2.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 5.9 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.3 | 8.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 2.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.3 | 2.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 1.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 0.3 | 1.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 5.7 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.3 | 5.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 3.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 1.5 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.3 | 1.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.3 | 16.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.3 | 1.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 2.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 | 5.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 3.5 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 1.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.3 | 7.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 0.8 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.3 | 10.2 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.3 | 1.6 | GO:0071051 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 | 4.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 7.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 2.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.3 | 1.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 7.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 10.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.3 | 3.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.3 | 16.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.3 | 5.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.3 | 15.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) |
| 0.3 | 1.5 | GO:0060732 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 2.9 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 4.9 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 1.9 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.2 | 1.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 3.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 5.5 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.2 | 0.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 4.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 3.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 6.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 7.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.2 | 1.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.2 | 2.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 5.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 7.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 1.7 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.2 | 0.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 24.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.2 | 8.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.2 | 3.7 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 1.4 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.2 | 2.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.2 | 0.8 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.2 | 3.6 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.2 | 2.5 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.2 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 2.0 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.2 | 0.5 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.2 | 1.5 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 7.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 1.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 0.5 | GO:1904683 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) regulation of metalloendopeptidase activity(GO:1904683) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.2 | 16.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 6.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 1.7 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.2 | 5.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 4.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 0.6 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.2 | 2.7 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 2.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 1.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 0.4 | GO:1903925 | response to bisphenol A(GO:1903925) |
| 0.1 | 0.4 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 1.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 2.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 8.0 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 1.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 5.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 2.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.7 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 2.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.9 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 1.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.5 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 1.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.5 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 5.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 1.8 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 2.7 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 2.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 3.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.9 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 5.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 3.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 3.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 4.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 1.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 2.9 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.1 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 1.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 10.5 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.1 | 0.3 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 5.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 2.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 2.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 2.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.7 | GO:0010332 | response to gamma radiation(GO:0010332) |
| 0.1 | 0.4 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 1.2 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) |
| 0.1 | 0.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 2.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 4.4 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 0.5 | GO:1990001 | positive regulation of protein polyubiquitination(GO:1902916) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 0.4 | GO:0035745 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 6.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 1.2 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.1 | 2.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 2.5 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 4.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 9.3 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 2.9 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.7 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 4.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 5.7 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.0 | 22.9 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 5.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 3.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 1.4 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.5 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 22.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 11.1 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.4 | GO:1902117 | positive regulation of organelle assembly(GO:1902117) |
| 0.0 | 0.2 | GO:0008542 | visual learning(GO:0008542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 15.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 3.4 | 10.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 3.4 | 10.2 | GO:0070821 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 2.6 | 13.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 2.6 | 10.3 | GO:0044393 | microspike(GO:0044393) |
| 2.3 | 6.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 2.2 | 6.5 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 2.1 | 36.5 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 2.1 | 6.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 2.0 | 10.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.0 | 6.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 2.0 | 25.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 2.0 | 17.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.8 | 5.3 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 1.7 | 9.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.6 | 4.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.5 | 6.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.4 | 5.4 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 1.2 | 8.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.2 | 4.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.2 | 11.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.1 | 11.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.0 | 3.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.0 | 5.9 | GO:1990393 | 3M complex(GO:1990393) |
| 1.0 | 15.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.9 | 4.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.9 | 26.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.9 | 3.5 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.8 | 2.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.8 | 10.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.8 | 7.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.8 | 6.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.8 | 2.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.7 | 15.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.7 | 13.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.7 | 12.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.7 | 5.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 18.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.7 | 5.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.6 | 7.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.6 | 32.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.6 | 1.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.6 | 2.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.6 | 41.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.6 | 8.0 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.6 | 3.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.5 | 4.9 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 6.8 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.5 | 13.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.5 | 9.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 6.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.5 | 3.5 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.5 | 1.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.5 | 3.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.5 | 1.9 | GO:0031417 | NatC complex(GO:0031417) |
| 0.5 | 3.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 9.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 2.7 | GO:0044308 | axonal spine(GO:0044308) |
| 0.4 | 3.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 12.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 10.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 12.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 6.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.4 | 50.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.4 | 37.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.4 | 16.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 4.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 3.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 9.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 33.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 2.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 1.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 6.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 4.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.3 | 7.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 25.0 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 35.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.3 | 3.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.3 | 1.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 7.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 3.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 1.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 1.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 1.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 11.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.2 | 17.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 22.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 8.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 5.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 13.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 11.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 1.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 4.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 2.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 4.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 11.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 7.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 1.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 5.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 2.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 3.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 5.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 42.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 11.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 6.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 13.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 2.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 2.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 6.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 6.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 3.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 4.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 5.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 2.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 28.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 4.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 5.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 10.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 4.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 3.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.7 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.5 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 15.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 18.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 3.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.3 | GO:0098800 | inner mitochondrial membrane protein complex(GO:0098800) |
| 0.1 | 1.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 9.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 9.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 30.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 8.5 | 25.6 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 5.4 | 26.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 4.6 | 22.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.9 | 19.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 3.3 | 9.9 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 2.8 | 8.5 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 2.6 | 10.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 2.5 | 7.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.0 | 9.9 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.8 | 7.3 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.7 | 7.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.7 | 15.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 1.7 | 5.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 1.5 | 17.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 1.5 | 7.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.4 | 5.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.4 | 10.1 | GO:0043426 | MRF binding(GO:0043426) |
| 1.4 | 5.7 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.4 | 5.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.4 | 8.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 1.4 | 5.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 1.3 | 6.7 | GO:0032767 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 1.3 | 5.3 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 1.3 | 11.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.3 | 6.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.2 | 3.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.2 | 4.9 | GO:0019002 | GMP binding(GO:0019002) |
| 1.2 | 3.5 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 1.1 | 6.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.1 | 4.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.1 | 5.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.0 | 8.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.0 | 7.9 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 1.0 | 7.7 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 1.0 | 42.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.9 | 4.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.9 | 3.5 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.8 | 14.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.8 | 9.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.8 | 2.3 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.8 | 3.8 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.8 | 5.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.8 | 9.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.7 | 8.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.7 | 5.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.7 | 2.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.7 | 45.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.7 | 4.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.7 | 6.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.7 | 2.0 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.7 | 8.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 3.3 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.6 | 3.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.6 | 16.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 3.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.6 | 4.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 4.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.6 | 10.7 | GO:0005523 | tropomyosin binding(GO:0005523) filamin binding(GO:0031005) |
| 0.6 | 4.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.6 | 4.5 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.6 | 1.7 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.6 | 1.7 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 3.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.5 | 7.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 6.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.5 | 3.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.5 | 9.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 4.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.5 | 4.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 2.7 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.5 | 3.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 4.7 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.5 | 3.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.5 | 2.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 3.0 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.5 | 28.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 11.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.5 | 4.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.5 | 15.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.5 | 7.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.5 | 9.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.5 | 3.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.5 | 2.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.5 | 7.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 7.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 19.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.4 | 1.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 9.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 5.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 6.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.4 | 8.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.4 | 6.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 7.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 3.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.3 | 2.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 8.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 2.9 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.3 | 7.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 3.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.3 | 1.5 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.3 | 5.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 28.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.3 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 2.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 2.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 3.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 3.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.3 | 12.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 1.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 4.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 4.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 29.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 1.7 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.2 | 6.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 0.7 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 64.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 25.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 8.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 1.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 15.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.2 | 67.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 51.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 3.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 4.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 6.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 1.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 9.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 6.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 3.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 2.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 5.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 1.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 14.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 2.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 33.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 0.7 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.2 | 4.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 41.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 0.8 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 5.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 1.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 2.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.2 | 2.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 1.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 5.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.7 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.1 | 1.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 2.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 4.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 5.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 6.4 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.1 | 4.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 40.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 1.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 7.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 10.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 11.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 5.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 11.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 6.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 3.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.6 | GO:0003916 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type I activity(GO:0003917) supercoiled DNA binding(GO:0097100) |
| 0.1 | 22.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 4.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 14.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 4.4 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 5.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 5.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 3.7 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 2.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.8 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 5.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 37.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.5 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 1.1 | GO:0008047 | enzyme activator activity(GO:0008047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 1.2 | 27.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 1.1 | 44.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.8 | 22.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.8 | 35.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.7 | 3.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.7 | 35.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.5 | 24.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.5 | 42.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.5 | 15.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.5 | 22.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.5 | 27.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.5 | 20.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.4 | 7.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.4 | 7.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 6.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 11.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 6.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.4 | 4.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 15.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.4 | 3.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 9.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.4 | 19.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.4 | 15.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.3 | 12.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.3 | 10.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 11.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 3.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 4.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 2.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 3.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 15.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 11.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 9.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 3.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 9.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 9.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 5.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 7.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 11.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 3.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 7.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 6.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 2.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 20.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 4.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 21.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 59.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.6 | 22.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.4 | 19.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 1.3 | 7.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.1 | 12.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 1.0 | 15.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.9 | 24.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.8 | 7.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.8 | 7.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.8 | 15.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.8 | 10.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 9.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.7 | 5.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.7 | 10.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.7 | 28.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.7 | 11.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.7 | 8.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.7 | 6.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.6 | 7.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.6 | 6.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.6 | 5.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.6 | 4.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.6 | 30.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.6 | 5.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 13.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.5 | 9.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.5 | 10.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.5 | 8.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.4 | 3.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 6.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 9.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 6.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.4 | 11.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 4.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 8.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.4 | 9.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 37.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.3 | 8.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 10.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.3 | 3.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 5.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 3.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 3.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 8.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 4.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 3.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 2.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 23.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 5.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 10.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 6.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 4.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 11.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 6.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 22.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 8.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 14.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 6.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 2.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 2.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 2.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 3.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 3.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 8.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 5.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 7.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 2.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 6.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 4.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 6.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 3.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 2.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 15.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 5.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 6.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.7 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 4.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 4.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |